Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for PAX1_PAX9
Z-value: 3.92
Transcription factors associated with PAX1_PAX9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
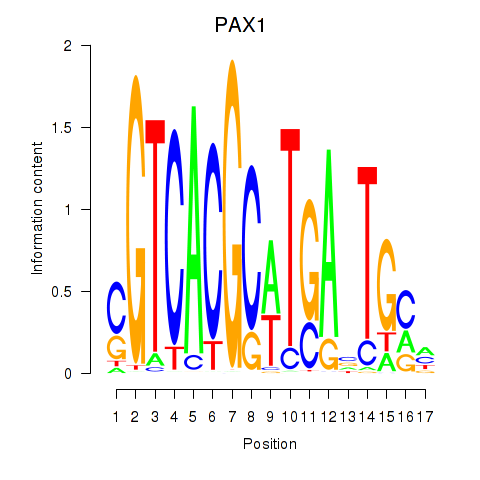
PAX1
|
ENSG00000125813.14 | PAX1 |
|
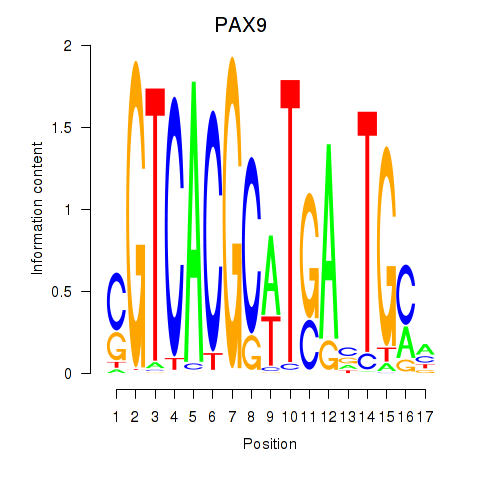
PAX9
|
ENSG00000198807.13 | PAX9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX9 | hg38_v1_chr14_+_36661852_36661939, hg38_v1_chr14_+_36657560_36657594 | -0.41 | 1.8e-10 | Click! |
| PAX1 | hg38_v1_chr20_+_21705651_21705673 | -0.40 | 5.3e-10 | Click! |
Activity profile of PAX1_PAX9 motif
Sorted Z-values of PAX1_PAX9 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX1_PAX9
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 12.3 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 2.7 | 35.0 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 2.5 | 7.4 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 2.2 | 19.6 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 1.3 | 9.0 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 1.1 | 5.7 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 1.1 | 8.0 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 1.1 | 8.0 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.6 | 1.8 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.6 | 2.4 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.5 | 7.0 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.4 | 6.2 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.4 | 3.0 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.3 | 17.4 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.3 | 8.0 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.2 | 2.0 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.2 | 32.9 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.2 | 2.7 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.2 | 15.2 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.1 | 14.0 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.1 | 2.2 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.1 | 2.9 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 10.5 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.8 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 2.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 3.5 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 2.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.3 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.7 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 32.5 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 2.1 | 10.5 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.8 | 8.0 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.7 | 3.0 | GO:0000811 | GINS complex(GO:0000811) |
| 0.4 | 41.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.2 | 12.3 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.2 | 35.0 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 15.2 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 7.0 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.1 | 8.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 2.2 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 2.9 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 3.5 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 2.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 5.7 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 7.4 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 2.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 17.0 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 32.5 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 3.5 | 17.4 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 2.7 | 8.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 2.5 | 7.4 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 0.6 | 5.7 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.6 | 2.4 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.4 | 15.2 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.3 | 8.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.3 | 1.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.3 | 1.8 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.2 | 8.0 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.2 | 3.0 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.2 | 7.0 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.2 | 2.1 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.2 | 12.3 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 9.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 35.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 23.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 2.4 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 6.2 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 1.3 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 3.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 35.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.3 | 7.0 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 9.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 8.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 20.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 35.0 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.4 | 8.0 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.3 | 64.2 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.3 | 7.0 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.3 | 8.0 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.2 | 9.0 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.2 | 3.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 3.5 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.1 | 17.3 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 0.7 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 5.7 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.1 | 4.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |