Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for PAX7_NOBOX
Z-value: 5.26
Transcription factors associated with PAX7_NOBOX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
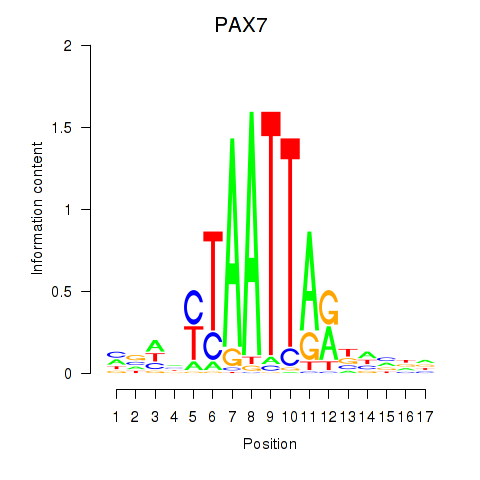
PAX7
|
ENSG00000009709.12 | PAX7 |
|
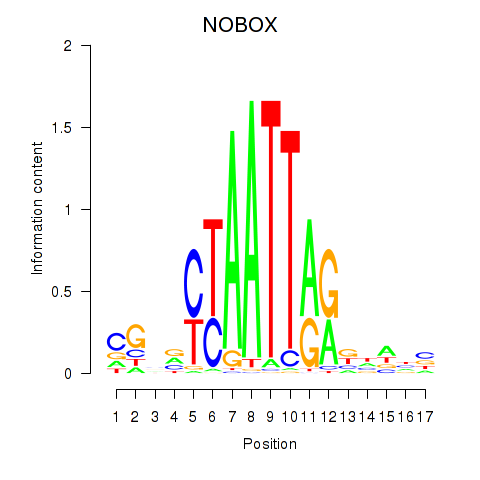
NOBOX
|
ENSG00000106410.15 | NOBOX |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX7 | hg38_v1_chr1_+_18631513_18631529 | 0.48 | 5.8e-14 | Click! |
Activity profile of PAX7_NOBOX motif
Sorted Z-values of PAX7_NOBOX motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX7_NOBOX
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.4 | 22.1 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 4.7 | 23.5 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 4.6 | 27.5 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 3.0 | 9.0 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 2.9 | 20.2 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 2.6 | 7.8 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 2.6 | 7.8 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 2.0 | 6.1 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 2.0 | 25.5 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 1.8 | 5.5 | GO:1900114 | positive regulation of histone H3-K9 trimethylation(GO:1900114) |
| 1.7 | 5.0 | GO:0071139 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 1.6 | 8.1 | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 1.6 | 6.2 | GO:0035627 | ceramide transport(GO:0035627) |
| 1.5 | 4.6 | GO:0052331 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 1.5 | 4.4 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 1.3 | 5.0 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 1.2 | 4.7 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 1.2 | 3.5 | GO:1904742 | protein auto-ADP-ribosylation(GO:0070213) regulation of telomeric DNA binding(GO:1904742) |
| 1.1 | 4.4 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) urea transmembrane transport(GO:0071918) purine nucleobase transmembrane transport(GO:1904823) |
| 1.1 | 3.2 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 1.0 | 6.8 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 1.0 | 2.9 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.9 | 3.7 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.9 | 11.6 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.8 | 2.5 | GO:0015847 | putrescine transport(GO:0015847) |
| 0.8 | 5.0 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.8 | 5.8 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.8 | 11.4 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.8 | 3.2 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.8 | 4.7 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.7 | 2.2 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.7 | 3.0 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.7 | 2.8 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.6 | 1.3 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.6 | 1.9 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.6 | 3.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.6 | 4.4 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.6 | 5.0 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.6 | 1.8 | GO:1904432 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.6 | 9.4 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.6 | 2.9 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.6 | 19.1 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.6 | 1.7 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.6 | 2.3 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.6 | 1.7 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.6 | 11.7 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.5 | 1.6 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.5 | 8.5 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.5 | 1.6 | GO:0002668 | negative regulation of T cell tolerance induction(GO:0002665) negative regulation of T cell anergy(GO:0002668) negative regulation of lymphocyte anergy(GO:0002912) regulation of lymphotoxin A production(GO:0032681) positive regulation of lymphotoxin A production(GO:0032761) regulation of lymphotoxin A biosynthetic process(GO:0043016) positive regulation of lymphotoxin A biosynthetic process(GO:0043017) |
| 0.5 | 3.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.5 | 2.0 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.5 | 1.5 | GO:0070055 | mRNA splicing via endonucleolytic cleavage and ligation involved in unfolded protein response(GO:0030969) mRNA splicing, via endonucleolytic cleavage and ligation(GO:0070054) mRNA endonucleolytic cleavage involved in unfolded protein response(GO:0070055) |
| 0.5 | 3.5 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.5 | 2.4 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.5 | 14.3 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.5 | 6.6 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.5 | 2.3 | GO:0097283 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.5 | 10.6 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.5 | 2.8 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.5 | 4.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) positive regulation of viral entry into host cell(GO:0046598) |
| 0.5 | 4.1 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.4 | 4.0 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.4 | 9.8 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.4 | 3.8 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.4 | 0.8 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.4 | 1.2 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.4 | 8.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.4 | 4.4 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.4 | 2.8 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.4 | 1.2 | GO:1902809 | skeletal muscle fiber differentiation(GO:0098528) regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.4 | 0.4 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.4 | 0.7 | GO:0002428 | antigen processing and presentation of peptide antigen via MHC class Ib(GO:0002428) |
| 0.3 | 2.7 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.3 | 13.3 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.3 | 13.6 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.3 | 60.4 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.3 | 2.6 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.3 | 2.0 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.3 | 7.5 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.3 | 0.6 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.3 | 2.6 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.3 | 3.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.3 | 7.1 | GO:0044126 | regulation of growth of symbiont in host(GO:0044126) |
| 0.3 | 4.6 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.3 | 0.3 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.3 | 8.4 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.3 | 1.2 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.3 | 1.7 | GO:0014004 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.3 | 3.9 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.3 | 3.6 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.3 | 0.8 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.3 | 4.3 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.3 | 1.3 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.3 | 2.1 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.3 | 0.8 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.2 | 1.0 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.2 | 0.7 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.2 | 1.5 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.2 | 6.4 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.2 | 1.4 | GO:0050955 | thermoception(GO:0050955) |
| 0.2 | 0.9 | GO:0045658 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.2 | 6.7 | GO:0036314 | response to sterol(GO:0036314) |
| 0.2 | 0.9 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.2 | 1.1 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.2 | 2.5 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.2 | 2.8 | GO:0098856 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.2 | 3.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.2 | 2.0 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.2 | 6.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.2 | 3.5 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.2 | 1.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.2 | 2.8 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.2 | 2.3 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.2 | 6.9 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.2 | 1.4 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.2 | 5.6 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.2 | 0.7 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.2 | 5.1 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.2 | 0.5 | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) |
| 0.2 | 3.5 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.2 | 3.0 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.2 | 1.8 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.1 | 2.1 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 0.6 | GO:0006581 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.1 | 1.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 1.4 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.1 | 0.7 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.1 | 4.4 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 1.0 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 11.8 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 0.9 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.1 | 3.4 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 1.7 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 3.0 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 0.1 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.1 | 1.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.8 | GO:0048840 | otolith development(GO:0048840) |
| 0.1 | 2.2 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 0.9 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.1 | 4.2 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 0.6 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 3.0 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 1.8 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.1 | 10.5 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 3.5 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.1 | 0.7 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.6 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 5.8 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.1 | 0.9 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 1.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.1 | 2.5 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.5 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.1 | 3.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 1.6 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 1.0 | GO:0008595 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.1 | 0.6 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 5.1 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.1 | 33.2 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.1 | 0.3 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.6 | GO:0002820 | negative regulation of adaptive immune response(GO:0002820) |
| 0.1 | 0.4 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 1.4 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.1 | 1.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.7 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 6.7 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.1 | 4.3 | GO:0002456 | T cell mediated immunity(GO:0002456) |
| 0.1 | 1.8 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 3.3 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.1 | 6.0 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 0.2 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.8 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 4.4 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 1.2 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 0.1 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.0 | 1.2 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 1.2 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.0 | 2.6 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.6 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 | 6.8 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 2.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 2.0 | GO:0050918 | positive chemotaxis(GO:0050918) |
| 0.0 | 0.7 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 1.7 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 4.0 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 4.9 | GO:0007272 | ensheathment of neurons(GO:0007272) axon ensheathment(GO:0008366) |
| 0.0 | 0.8 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.9 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 2.6 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.7 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.5 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 1.0 | GO:0031640 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.0 | 2.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 1.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.6 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 0.7 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 1.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 4.7 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 1.4 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 3.3 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.4 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 1.6 | GO:0000725 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 0.6 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 1.2 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 1.9 | GO:0030324 | lung development(GO:0030324) |
| 0.0 | 1.5 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 0.8 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.2 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.0 | 0.6 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.5 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 7.0 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 2.1 | 6.4 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 2.0 | 25.5 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 1.4 | 4.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 1.4 | 8.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 1.2 | 8.5 | GO:0060091 | kinocilium(GO:0060091) |
| 1.1 | 11.9 | GO:0043203 | axon hillock(GO:0043203) |
| 1.0 | 13.6 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 1.0 | 8.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 1.0 | 11.7 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.9 | 2.7 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.9 | 6.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.8 | 5.0 | GO:0033063 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.8 | 2.4 | GO:0043159 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) acrosomal matrix(GO:0043159) |
| 0.8 | 4.7 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.7 | 11.7 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.7 | 3.6 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.7 | 2.8 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.7 | 10.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.7 | 8.6 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.6 | 2.6 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.6 | 5.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.6 | 4.4 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.5 | 3.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.5 | 3.7 | GO:0070187 | telosome(GO:0070187) |
| 0.5 | 1.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.4 | 16.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.4 | 4.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.4 | 1.6 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.4 | 5.9 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.4 | 1.8 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.4 | 3.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.3 | 3.7 | GO:0005638 | lamin filament(GO:0005638) |
| 0.3 | 8.7 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.3 | 3.5 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.3 | 1.8 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.3 | 3.8 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.2 | 1.0 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.2 | 3.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.2 | 7.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 5.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 4.7 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.2 | 1.6 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.2 | 3.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.2 | 2.0 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.2 | 3.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.2 | 22.0 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 0.7 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 2.5 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 14.0 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 2.9 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 1.9 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 11.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 3.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 3.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 6.7 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 1.9 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 8.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 14.5 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 4.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.6 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 1.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 3.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.2 | GO:0036019 | endolysosome(GO:0036019) |
| 0.1 | 11.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 0.2 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.1 | 83.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 8.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.6 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 1.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 6.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.6 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.7 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 1.1 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 8.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 2.9 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 9.6 | GO:0005774 | vacuolar membrane(GO:0005774) |
| 0.0 | 1.3 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 3.4 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 15.1 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 2.0 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 1.0 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 20.9 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.9 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 22.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 3.9 | 27.5 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 3.0 | 18.3 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 2.0 | 9.9 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 1.6 | 6.2 | GO:0097001 | ceramide binding(GO:0097001) |
| 1.5 | 4.4 | GO:0015265 | urea channel activity(GO:0015265) |
| 1.3 | 8.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 1.1 | 3.4 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 1.1 | 4.4 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 1.0 | 3.1 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 1.0 | 5.9 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) |
| 0.9 | 4.4 | GO:0047374 | sterol esterase activity(GO:0004771) methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.9 | 12.8 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.8 | 2.5 | GO:0015203 | polyamine transmembrane transporter activity(GO:0015203) putrescine transmembrane transporter activity(GO:0015489) |
| 0.8 | 5.9 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.8 | 2.5 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.8 | 4.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.8 | 8.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.8 | 3.2 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.7 | 3.0 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.7 | 2.9 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.7 | 4.3 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.7 | 5.0 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.7 | 2.8 | GO:0035276 | aldehyde oxidase activity(GO:0004031) ethanol binding(GO:0035276) |
| 0.7 | 2.1 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.7 | 2.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.6 | 6.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.6 | 2.6 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.6 | 1.9 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.6 | 5.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.6 | 1.9 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.6 | 5.5 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.6 | 1.8 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.6 | 3.5 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.6 | 6.8 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.5 | 1.6 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.5 | 4.7 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.5 | 2.8 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.4 | 5.8 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.4 | 7.6 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.4 | 1.2 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.4 | 5.8 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.4 | 49.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.4 | 11.7 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.4 | 3.2 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.4 | 1.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.4 | 8.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.4 | 2.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.4 | 23.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.4 | 3.6 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.4 | 5.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.3 | 4.9 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.3 | 7.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.3 | 6.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.3 | 12.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.3 | 33.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.3 | 5.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.3 | 4.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.3 | 13.6 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.3 | 0.9 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.3 | 3.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.3 | 0.8 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.2 | 0.7 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.2 | 0.7 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.2 | 0.7 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.2 | 5.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 0.9 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.2 | 2.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.2 | 0.9 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.2 | 1.7 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 45.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.2 | 12.0 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.2 | 2.5 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.2 | 1.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.2 | 3.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.2 | 3.0 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.2 | 0.9 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.2 | 2.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.2 | 1.4 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.2 | 3.7 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 14.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.2 | 0.7 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.2 | 2.9 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.4 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 1.7 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 1.6 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 1.4 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 0.6 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.1 | 1.8 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 10.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 2.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.8 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 3.0 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 1.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 37.0 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 0.9 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 0.6 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 20.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 3.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.3 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 0.9 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 4.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 1.5 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 1.1 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 1.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.7 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 1.6 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.3 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.1 | 1.8 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.1 | 0.7 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.1 | 0.6 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.1 | 3.7 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 3.4 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.1 | 2.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 2.2 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 0.7 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.1 | 0.2 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.1 | 6.0 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 10.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 2.0 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.6 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 1.1 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.1 | 4.3 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 3.5 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 1.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 12.0 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.6 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 4.7 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 3.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 4.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.6 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 2.3 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 11.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 1.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 9.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 1.6 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.2 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 1.7 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 1.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.5 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.3 | GO:0050136 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 7.3 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 1.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 2.4 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.7 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 2.3 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 12.2 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.4 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 1.2 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 1.7 | GO:0005178 | integrin binding(GO:0005178) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 23.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.5 | 21.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.3 | 16.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.3 | 8.7 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.2 | 16.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 60.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 12.7 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.2 | 4.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 7.0 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 9.8 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 8.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 8.2 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 2.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 1.0 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 15.6 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 1.1 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.1 | 1.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 3.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 4.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 9.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 4.4 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 4.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 2.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 2.4 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 1.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 10.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 0.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 0.9 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 1.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 2.5 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 7.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.9 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.7 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 3.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 2.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 13.6 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.9 | 25.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.6 | 6.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.5 | 9.9 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.5 | 11.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.5 | 11.7 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.5 | 3.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.4 | 6.5 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.4 | 9.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.4 | 8.3 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.3 | 4.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 3.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.3 | 2.8 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.3 | 7.3 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.3 | 12.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.3 | 3.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 2.5 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.2 | 11.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.2 | 3.9 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.2 | 9.5 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.2 | 9.0 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 3.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 7.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.2 | 4.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 1.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.2 | 5.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 6.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.2 | 29.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 4.1 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 8.2 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 2.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 3.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 4.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 3.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 1.9 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 2.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.6 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 2.0 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 3.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 4.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 3.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 1.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 1.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 6.7 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 2.6 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 2.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 5.8 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 1.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 2.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 2.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 22.8 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.1 | 2.0 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.1 | 1.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 1.1 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 2.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 1.6 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 1.0 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.2 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 1.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 1.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.3 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 1.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.8 | REACTOME SIGNALING BY TGF BETA RECEPTOR COMPLEX | Genes involved in Signaling by TGF-beta Receptor Complex |
| 0.0 | 1.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.7 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 1.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.5 | REACTOME UNFOLDED PROTEIN RESPONSE | Genes involved in Unfolded Protein Response |
| 0.0 | 0.9 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.6 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 1.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.2 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 1.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |