Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
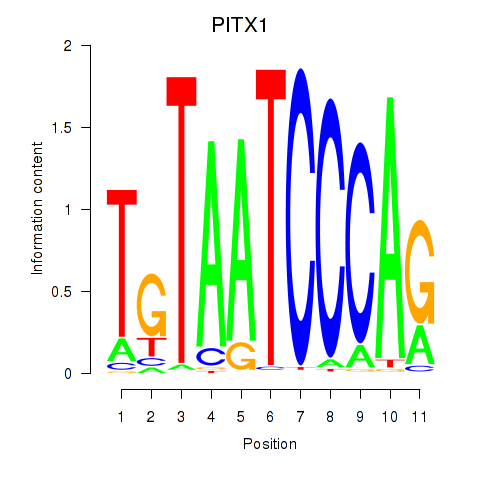
Results for PITX1
Z-value: 8.31
Transcription factors associated with PITX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PITX1
|
ENSG00000069011.16 | PITX1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PITX1 | hg38_v1_chr5_-_135034212_135034294 | -0.00 | 9.9e-01 | Click! |
Activity profile of PITX1 motif
Sorted Z-values of PITX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PITX1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.4 | 34.1 | GO:0072019 | proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 11.1 | 77.9 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 11.0 | 32.9 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 9.4 | 28.3 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 9.2 | 73.3 | GO:0051343 | positive regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051343) |
| 9.0 | 36.1 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 8.2 | 106.7 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 8.1 | 32.4 | GO:0043060 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 7.4 | 44.2 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 7.3 | 22.0 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 7.0 | 70.1 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 6.5 | 45.8 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 6.3 | 31.3 | GO:0046103 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 6.2 | 24.9 | GO:0003290 | atrial septum secundum morphogenesis(GO:0003290) |
| 5.9 | 35.7 | GO:0044211 | CTP salvage(GO:0044211) |
| 5.6 | 33.5 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 5.5 | 22.0 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 5.5 | 32.8 | GO:1900369 | regulation of RNA interference(GO:1900368) negative regulation of RNA interference(GO:1900369) |
| 5.5 | 21.8 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 5.2 | 15.6 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 5.1 | 20.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 5.0 | 15.0 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 4.9 | 9.7 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 4.8 | 33.5 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 4.8 | 14.3 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 4.7 | 9.4 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 4.6 | 13.9 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 4.5 | 59.0 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 4.4 | 17.7 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 4.3 | 38.9 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 4.3 | 4.3 | GO:0046984 | regulation of hemoglobin biosynthetic process(GO:0046984) |
| 4.2 | 16.9 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 4.2 | 12.7 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 4.2 | 92.6 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 4.2 | 46.3 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 4.2 | 25.2 | GO:1990539 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 4.2 | 33.3 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 4.1 | 12.4 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 4.1 | 32.5 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 4.0 | 11.9 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 3.9 | 43.4 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 3.8 | 19.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 3.8 | 26.3 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 3.7 | 40.7 | GO:1901977 | negative regulation of cell cycle checkpoint(GO:1901977) |
| 3.6 | 10.9 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 3.5 | 31.9 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 3.5 | 31.3 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 3.5 | 20.8 | GO:2001184 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 3.4 | 23.8 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 3.4 | 13.5 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 3.3 | 13.3 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 3.2 | 9.6 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 3.2 | 9.5 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 3.2 | 6.3 | GO:0071035 | polyadenylation-dependent ncRNA catabolic process(GO:0043634) nuclear ncRNA surveillance(GO:0071029) nuclear polyadenylation-dependent rRNA catabolic process(GO:0071035) nuclear polyadenylation-dependent ncRNA catabolic process(GO:0071046) |
| 3.1 | 6.3 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 3.1 | 34.4 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 3.1 | 9.3 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 3.1 | 6.2 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 3.0 | 33.5 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 3.0 | 18.0 | GO:0016036 | cellular response to phosphate starvation(GO:0016036) positive regulation of sulfur amino acid metabolic process(GO:0031337) positive regulation of homocysteine metabolic process(GO:0050668) |
| 3.0 | 12.0 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 2.9 | 123.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 2.9 | 23.1 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 2.8 | 11.1 | GO:0071879 | UDP-glucose catabolic process(GO:0006258) positive regulation of adrenergic receptor signaling pathway(GO:0071879) negative regulation of type B pancreatic cell development(GO:2000077) |
| 2.8 | 41.3 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 2.7 | 19.0 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 2.7 | 8.1 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 2.7 | 37.3 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 2.6 | 7.8 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 2.6 | 10.3 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 2.6 | 5.1 | GO:0014806 | smooth muscle hyperplasia(GO:0014806) |
| 2.6 | 20.4 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 2.5 | 5.1 | GO:0032262 | pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) |
| 2.5 | 7.6 | GO:0002339 | B cell selection(GO:0002339) |
| 2.5 | 7.5 | GO:0036233 | glycine import(GO:0036233) |
| 2.5 | 94.4 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 2.5 | 14.9 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 2.5 | 12.4 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 2.5 | 52.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 2.4 | 9.5 | GO:0070981 | L-asparagine biosynthetic process(GO:0070981) L-asparagine metabolic process(GO:0070982) |
| 2.3 | 9.4 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 2.3 | 11.5 | GO:0045337 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 2.3 | 13.8 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 2.2 | 36.0 | GO:0043248 | proteasome assembly(GO:0043248) |
| 2.2 | 28.9 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 2.2 | 8.8 | GO:0035803 | egg coat formation(GO:0035803) |
| 2.2 | 6.6 | GO:0035915 | pore formation in membrane of other organism(GO:0035915) |
| 2.2 | 41.4 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 2.2 | 12.9 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) |
| 2.1 | 18.8 | GO:0007070 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 2.1 | 6.2 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 2.0 | 2.0 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 2.0 | 18.0 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 2.0 | 3.9 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 1.9 | 73.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 1.9 | 13.4 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 1.9 | 158.6 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 1.9 | 9.3 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 1.9 | 9.3 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 1.8 | 9.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 1.8 | 5.5 | GO:0034402 | mRNA export from nucleus in response to heat stress(GO:0031990) recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 1.8 | 20.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 1.8 | 59.8 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 1.8 | 10.9 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 1.8 | 7.2 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 1.8 | 82.0 | GO:0000303 | response to superoxide(GO:0000303) |
| 1.8 | 3.6 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 1.8 | 10.5 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 1.7 | 1.7 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 1.7 | 8.4 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 1.7 | 16.8 | GO:0046689 | response to mercury ion(GO:0046689) |
| 1.7 | 5.0 | GO:2001187 | positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 1.6 | 35.4 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 1.6 | 26.9 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 1.6 | 15.6 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 1.6 | 18.8 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 1.5 | 86.8 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 1.5 | 1.5 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 1.5 | 6.2 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 1.5 | 10.8 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 1.5 | 111.7 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 1.5 | 7.6 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 1.5 | 6.1 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 1.5 | 3.0 | GO:0010899 | regulation of phosphatidylcholine catabolic process(GO:0010899) |
| 1.5 | 20.9 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) |
| 1.5 | 8.9 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 1.5 | 4.4 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 1.5 | 13.2 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 1.5 | 5.8 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 1.4 | 29.0 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 1.4 | 8.7 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 1.4 | 4.3 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 1.4 | 11.1 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 1.4 | 13.9 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 1.4 | 5.5 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 1.4 | 9.5 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 1.3 | 6.6 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 1.3 | 11.9 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 1.3 | 21.1 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 1.3 | 15.7 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 1.3 | 10.4 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 1.3 | 18.0 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 1.3 | 45.0 | GO:0034080 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 1.3 | 111.8 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 1.3 | 10.1 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 1.3 | 17.6 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 1.2 | 17.5 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 1.2 | 6.0 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 1.2 | 3.6 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 1.2 | 2.4 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 1.2 | 6.0 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 1.2 | 109.5 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 1.2 | 3.5 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 1.2 | 18.5 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 1.1 | 3.4 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 1.1 | 3.4 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 1.1 | 6.8 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 1.1 | 5.6 | GO:2001107 | negative regulation of establishment of T cell polarity(GO:1903904) negative regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001107) |
| 1.1 | 20.1 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 1.1 | 7.8 | GO:0071501 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 1.1 | 6.6 | GO:0003051 | angiotensin catabolic process in blood(GO:0002005) angiotensin-mediated drinking behavior(GO:0003051) multicellular organismal protein catabolic process(GO:0044254) protein digestion(GO:0044256) multicellular organismal macromolecule catabolic process(GO:0044266) |
| 1.1 | 22.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 1.1 | 15.4 | GO:0035581 | sequestering of extracellular ligand from receptor(GO:0035581) |
| 1.1 | 6.5 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 1.1 | 9.5 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 1.1 | 6.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 1.0 | 5.2 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 1.0 | 4.1 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 1.0 | 2.0 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 1.0 | 26.9 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 1.0 | 3.0 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 1.0 | 12.8 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 1.0 | 13.7 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 1.0 | 56.4 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.9 | 65.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.9 | 11.1 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.9 | 3.7 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.9 | 4.5 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.9 | 12.5 | GO:0043486 | histone exchange(GO:0043486) |
| 0.9 | 34.7 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.9 | 8.5 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.8 | 11.0 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.8 | 1.6 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.8 | 1.6 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.8 | 18.7 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.8 | 18.5 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.8 | 7.2 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) |
| 0.8 | 19.9 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.8 | 7.1 | GO:0045047 | protein targeting to ER(GO:0045047) |
| 0.8 | 4.0 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.8 | 2.4 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.8 | 3.9 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.8 | 3.9 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.8 | 20.8 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.8 | 1.5 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.8 | 15.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.8 | 1.5 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.8 | 5.3 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.8 | 27.3 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.8 | 6.8 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.7 | 3.7 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.7 | 3.0 | GO:1903899 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.7 | 5.9 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.7 | 7.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.7 | 51.9 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.7 | 3.6 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.7 | 9.4 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.7 | 4.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.7 | 16.3 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.7 | 4.0 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.7 | 63.4 | GO:0006413 | translational initiation(GO:0006413) |
| 0.7 | 3.3 | GO:0098502 | DNA dephosphorylation(GO:0098502) |
| 0.6 | 7.1 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.6 | 3.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.6 | 8.9 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.6 | 2.5 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.6 | 3.7 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.6 | 8.0 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.6 | 2.4 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.6 | 4.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.6 | 1.7 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.6 | 9.3 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.6 | 2.9 | GO:1901094 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.6 | 9.1 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.6 | 3.4 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 0.6 | 10.8 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.6 | 5.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.6 | 3.4 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.6 | 2.8 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.6 | 2.2 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.6 | 5.0 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.5 | 2.7 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.5 | 3.8 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.5 | 1.6 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.5 | 1.6 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.5 | 5.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.5 | 3.7 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.5 | 2.7 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) regulation of voltage-gated chloride channel activity(GO:1902941) positive regulation of voltage-gated chloride channel activity(GO:1902943) positive regulation of leukocyte tethering or rolling(GO:1903238) positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.5 | 5.8 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.5 | 1.6 | GO:0042704 | negative regulation of alkaline phosphatase activity(GO:0010693) uterine wall breakdown(GO:0042704) |
| 0.5 | 5.2 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.5 | 4.6 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.5 | 2.1 | GO:0070316 | regulation of G0 to G1 transition(GO:0070316) |
| 0.5 | 11.7 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.5 | 1.0 | GO:0033007 | negative regulation of mast cell activation involved in immune response(GO:0033007) |
| 0.5 | 8.7 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.5 | 1.4 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.5 | 0.5 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.5 | 13.8 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.5 | 7.5 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.5 | 8.3 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.5 | 6.4 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.5 | 43.6 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.4 | 1.3 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.4 | 5.3 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.4 | 18.0 | GO:0061615 | NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.4 | 23.7 | GO:0018208 | peptidyl-proline modification(GO:0018208) |
| 0.4 | 2.6 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.4 | 5.4 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.4 | 105.3 | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.4 | 0.4 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.4 | 1.6 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.4 | 25.9 | GO:0043303 | mast cell activation involved in immune response(GO:0002279) mast cell degranulation(GO:0043303) |
| 0.4 | 5.9 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.4 | 1.9 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.4 | 1.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.4 | 4.9 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.4 | 6.8 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.4 | 8.3 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.4 | 21.6 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.4 | 4.8 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.4 | 1.1 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.4 | 2.5 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.4 | 6.0 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.4 | 0.7 | GO:0006288 | base-excision repair, DNA ligation(GO:0006288) |
| 0.3 | 3.8 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.3 | 2.4 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.3 | 3.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.3 | 6.2 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.3 | 10.0 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.3 | 3.6 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.3 | 4.3 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.3 | 1.0 | GO:0051039 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.3 | 2.9 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.3 | 3.8 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.3 | 2.2 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.3 | 1.8 | GO:0014057 | positive regulation of glutamate secretion(GO:0014049) positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.3 | 6.1 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.3 | 9.6 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.3 | 6.0 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.3 | 4.4 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.3 | 7.3 | GO:0072663 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.3 | 1.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.3 | 10.9 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.3 | 2.2 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.3 | 1.4 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 0.3 | 0.8 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.3 | 15.3 | GO:0050850 | positive regulation of calcium-mediated signaling(GO:0050850) |
| 0.3 | 1.0 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.3 | 2.9 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.3 | 16.8 | GO:0035308 | negative regulation of protein dephosphorylation(GO:0035308) |
| 0.3 | 5.0 | GO:2000696 | regulation of epithelial cell differentiation involved in kidney development(GO:2000696) |
| 0.2 | 1.0 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.2 | 5.0 | GO:0006266 | DNA ligation(GO:0006266) |
| 0.2 | 2.0 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.2 | 9.3 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.2 | 0.9 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.2 | 14.7 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.2 | 1.4 | GO:1903911 | positive regulation of receptor clustering(GO:1903911) |
| 0.2 | 5.0 | GO:0006833 | water transport(GO:0006833) |
| 0.2 | 8.3 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.2 | 8.3 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.2 | 3.1 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.2 | 2.1 | GO:0010626 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.2 | 1.5 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.2 | 3.7 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.2 | 0.8 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.2 | 2.9 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.2 | 3.4 | GO:0044065 | regulation of respiratory system process(GO:0044065) |
| 0.2 | 1.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.2 | 8.6 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.2 | 1.4 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.2 | 1.8 | GO:0015871 | choline transport(GO:0015871) |
| 0.2 | 1.8 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.2 | 3.3 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.2 | 7.8 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.2 | 2.2 | GO:0032329 | serine transport(GO:0032329) |
| 0.2 | 0.5 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.2 | 9.6 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.2 | 1.9 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.2 | 0.8 | GO:1904646 | cellular response to beta-amyloid(GO:1904646) |
| 0.2 | 4.1 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.2 | 1.5 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 1.6 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 12.1 | GO:0034340 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 | 3.3 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 4.7 | GO:2001044 | regulation of integrin-mediated signaling pathway(GO:2001044) |
| 0.1 | 1.7 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 3.3 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 3.7 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.1 | 3.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 2.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 1.6 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 1.0 | GO:0006734 | NADH metabolic process(GO:0006734) |
| 0.1 | 1.6 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 4.0 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.1 | 1.6 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.1 | 3.1 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 14.0 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.1 | 0.3 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.1 | 9.3 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.1 | 2.8 | GO:0023058 | adaptation of signaling pathway(GO:0023058) |
| 0.1 | 4.9 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.1 | 2.1 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 10.5 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 4.1 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 1.9 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.1 | 4.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 8.3 | GO:0051149 | positive regulation of muscle cell differentiation(GO:0051149) |
| 0.1 | 1.7 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.1 | 2.9 | GO:0034383 | low-density lipoprotein particle clearance(GO:0034383) |
| 0.1 | 2.6 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 0.8 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.1 | 1.2 | GO:0051660 | cortical microtubule organization(GO:0043622) establishment of centrosome localization(GO:0051660) |
| 0.1 | 2.8 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 2.7 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 6.9 | GO:0008344 | adult locomotory behavior(GO:0008344) |
| 0.1 | 9.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 1.1 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 1.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 29.1 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.1 | 3.2 | GO:0009311 | oligosaccharide metabolic process(GO:0009311) |
| 0.1 | 0.8 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.5 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 1.8 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.1 | 0.4 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.1 | 0.3 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 0.8 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.1 | 2.8 | GO:0046427 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.1 | 3.2 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.1 | 1.0 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.1 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.0 | 0.3 | GO:0072319 | synaptic vesicle uncoating(GO:0016191) vesicle uncoating(GO:0072319) |
| 0.0 | 3.4 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.6 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.3 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.7 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 1.3 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.2 | GO:1902713 | regulation of interferon-gamma secretion(GO:1902713) positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 2.0 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.0 | 4.2 | GO:0002431 | Fc receptor mediated stimulatory signaling pathway(GO:0002431) |
| 0.0 | 1.1 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.3 | GO:0007077 | mitotic nuclear envelope disassembly(GO:0007077) |
| 0.0 | 0.7 | GO:0001662 | behavioral fear response(GO:0001662) behavioral defense response(GO:0002209) |
| 0.0 | 0.3 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 1.0 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.0 | GO:0060177 | regulation of angiotensin levels in blood(GO:0002002) angiotensin maturation(GO:0002003) drinking behavior(GO:0042756) regulation of angiotensin metabolic process(GO:0060177) |
| 0.0 | 0.2 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.6 | 31.7 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 10.5 | 73.8 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 10.2 | 91.7 | GO:0034709 | methylosome(GO:0034709) |
| 10.1 | 30.3 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 9.7 | 106.7 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 8.1 | 73.3 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 8.1 | 32.4 | GO:0005715 | late recombination nodule(GO:0005715) |
| 6.3 | 44.3 | GO:0005687 | U4 snRNP(GO:0005687) |
| 6.3 | 31.3 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 5.7 | 11.4 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 5.6 | 22.6 | GO:0071920 | cleavage body(GO:0071920) |
| 5.5 | 49.7 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 5.4 | 43.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 4.9 | 58.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 4.7 | 32.8 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 4.5 | 13.5 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 4.5 | 31.3 | GO:0032021 | NELF complex(GO:0032021) |
| 4.4 | 26.2 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 4.3 | 56.0 | GO:0042555 | MCM complex(GO:0042555) |
| 4.2 | 20.8 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 4.1 | 44.9 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 4.0 | 20.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 4.0 | 11.9 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 3.8 | 38.0 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 3.7 | 99.9 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 3.6 | 68.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 3.6 | 14.3 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 3.5 | 42.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 3.4 | 30.8 | GO:0070652 | HAUS complex(GO:0070652) |
| 3.4 | 23.8 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 3.4 | 10.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 3.3 | 52.8 | GO:0005642 | annulate lamellae(GO:0005642) |
| 3.2 | 29.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 3.1 | 6.2 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 3.1 | 18.5 | GO:0001940 | male pronucleus(GO:0001940) |
| 3.0 | 39.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 2.9 | 8.7 | GO:0055087 | Ski complex(GO:0055087) |
| 2.9 | 48.7 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 2.8 | 19.9 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 2.8 | 13.9 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 2.8 | 13.8 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 2.7 | 53.2 | GO:0005839 | proteasome core complex(GO:0005839) |
| 2.6 | 7.9 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 2.6 | 7.8 | GO:1990038 | glial cytoplasmic inclusion(GO:0097409) classical Lewy body(GO:0097414) Lewy neurite(GO:0097462) Lewy body corona(GO:1990038) |
| 2.6 | 20.9 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 2.5 | 20.4 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 2.5 | 10.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 2.4 | 60.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 2.4 | 42.7 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 2.3 | 11.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 2.2 | 11.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 2.2 | 23.9 | GO:0005638 | lamin filament(GO:0005638) |
| 2.2 | 6.5 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 2.1 | 31.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 2.1 | 64.3 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 2.1 | 195.9 | GO:0015935 | small ribosomal subunit(GO:0015935) |
| 2.1 | 10.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 2.0 | 28.7 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 1.9 | 17.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 1.9 | 11.1 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 1.9 | 14.8 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 1.9 | 25.9 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 1.7 | 15.7 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 1.7 | 13.3 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 1.7 | 16.7 | GO:0061574 | ASAP complex(GO:0061574) |
| 1.6 | 169.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 1.6 | 4.7 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 1.6 | 9.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 1.5 | 13.6 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 1.5 | 4.5 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 1.5 | 18.9 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 1.4 | 20.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 1.4 | 18.5 | GO:0070938 | contractile ring(GO:0070938) |
| 1.4 | 176.9 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 1.4 | 62.0 | GO:0000313 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 1.4 | 6.9 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 1.4 | 37.0 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 1.3 | 5.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 1.2 | 13.7 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 1.2 | 12.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 1.2 | 15.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 1.1 | 17.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 1.1 | 11.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 1.1 | 4.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 1.1 | 17.0 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 1.1 | 7.4 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 1.1 | 4.2 | GO:1990879 | CST complex(GO:1990879) |
| 1.0 | 12.6 | GO:0033177 | proton-transporting two-sector ATPase complex, proton-transporting domain(GO:0033177) proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 1.0 | 8.8 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.9 | 22.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.9 | 10.3 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.9 | 6.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.9 | 6.3 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.9 | 12.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.8 | 15.3 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.8 | 10.9 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.8 | 33.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.8 | 14.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.8 | 6.6 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.8 | 2.4 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.7 | 6.0 | GO:1905202 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.7 | 16.8 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.7 | 5.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.7 | 11.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.6 | 115.0 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.6 | 0.6 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.6 | 22.2 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.6 | 35.2 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.6 | 6.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.6 | 22.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.6 | 18.8 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.5 | 14.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.5 | 10.3 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.5 | 12.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.5 | 3.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.5 | 4.7 | GO:0044754 | autolysosome(GO:0044754) |
| 0.5 | 8.4 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.5 | 31.5 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.5 | 3.9 | GO:0030891 | VCB complex(GO:0030891) |
| 0.5 | 5.9 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.4 | 27.0 | GO:0030684 | preribosome(GO:0030684) |
| 0.4 | 5.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.4 | 34.4 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.4 | 4.4 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.4 | 18.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.4 | 1.5 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.4 | 1.2 | GO:0033011 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) |
| 0.4 | 2.7 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.4 | 4.3 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.3 | 48.2 | GO:0016605 | PML body(GO:0016605) |
| 0.3 | 39.5 | GO:0000922 | spindle pole(GO:0000922) |
| 0.3 | 1.4 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.3 | 5.9 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.3 | 5.8 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.3 | 28.3 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.3 | 3.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.3 | 5.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.3 | 3.8 | GO:0030681 | multimeric ribonuclease P complex(GO:0030681) |
| 0.3 | 3.0 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.3 | 6.0 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.3 | 3.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.3 | 17.0 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.3 | 15.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.3 | 3.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.3 | 3.9 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.3 | 3.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.3 | 1.6 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.3 | 14.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.3 | 6.7 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.3 | 4.6 | GO:0042611 | MHC protein complex(GO:0042611) MHC class II protein complex(GO:0042613) |
| 0.3 | 16.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.2 | 2.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.2 | 2.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.2 | 22.0 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.2 | 7.4 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.2 | 5.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 3.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 5.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.2 | 10.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 4.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 1.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 3.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.2 | 1.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.2 | 2.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 13.0 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 7.6 | GO:0032153 | cell division site(GO:0032153) cleavage furrow(GO:0032154) cell division site part(GO:0032155) cell surface furrow(GO:0097610) |
| 0.1 | 15.6 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 6.0 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.1 | 10.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 0.8 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 7.1 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.1 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 1.8 | GO:0036452 | ESCRT complex(GO:0036452) |
| 0.1 | 8.8 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 2.0 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 26.1 | GO:0030529 | intracellular ribonucleoprotein complex(GO:0030529) |
| 0.1 | 5.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 8.8 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 33.2 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 34.8 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 0.7 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.1 | 9.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 12.1 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 6.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 2.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 3.5 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.1 | 1.3 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 0.3 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 2.3 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.3 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.8 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 8.6 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 1.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.4 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 1.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 5.2 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.3 | GO:0097546 | ciliary base(GO:0097546) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.5 | 75.0 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 11.1 | 66.5 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 9.1 | 36.3 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 8.5 | 34.1 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 8.3 | 25.0 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 7.8 | 23.3 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 7.4 | 44.6 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 7.3 | 73.3 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 6.7 | 20.2 | GO:0003896 | DNA primase activity(GO:0003896) |
| 6.3 | 31.3 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 6.3 | 50.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 6.2 | 43.7 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 5.9 | 58.6 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 5.8 | 23.3 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 5.6 | 22.6 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 5.6 | 50.2 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 5.5 | 22.0 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 5.5 | 21.8 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 5.4 | 43.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 5.3 | 16.0 | GO:0080101 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 5.0 | 30.3 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 4.9 | 29.3 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 4.7 | 18.7 | GO:0043273 | CTPase activity(GO:0043273) |
| 4.6 | 18.6 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 4.6 | 32.4 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 4.5 | 17.9 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 4.5 | 44.5 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 4.3 | 17.3 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 4.2 | 16.9 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 4.2 | 25.2 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 4.2 | 33.3 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 4.1 | 32.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 3.8 | 84.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 3.8 | 60.4 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 3.8 | 56.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 3.7 | 14.9 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 3.6 | 14.3 | GO:0032357 | oxidized purine DNA binding(GO:0032357) |
| 3.5 | 31.9 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 3.5 | 62.6 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 3.2 | 9.6 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 3.1 | 28.3 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 3.1 | 9.4 | GO:0004750 | ribulose-phosphate 3-epimerase activity(GO:0004750) |
| 3.1 | 18.4 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 3.0 | 20.9 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 3.0 | 106.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 2.9 | 26.3 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 2.8 | 34.0 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 2.8 | 8.5 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 2.7 | 11.0 | GO:0002134 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 2.7 | 21.4 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 2.6 | 15.8 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 2.6 | 15.7 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 2.6 | 15.7 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 2.6 | 13.0 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 2.5 | 22.7 | GO:0032190 | acrosin binding(GO:0032190) |
| 2.5 | 42.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 2.4 | 67.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 2.3 | 7.0 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 2.3 | 11.5 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 2.2 | 13.2 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 2.2 | 58.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 2.1 | 8.3 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 2.0 | 9.9 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 1.9 | 84.8 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 1.9 | 13.4 | GO:0004459 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 1.9 | 9.5 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 1.9 | 20.8 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 1.8 | 12.8 | GO:0030614 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 1.8 | 5.4 | GO:0080023 | 3R-hydroxyacyl-CoA dehydratase activity(GO:0080023) |
| 1.8 | 8.9 | GO:0001129 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 1.7 | 8.7 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 1.7 | 6.9 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 1.7 | 5.1 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 1.7 | 10.3 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 1.7 | 13.7 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 1.7 | 13.4 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 1.7 | 340.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 1.7 | 5.0 | GO:0033699 | DNA 5'-adenosine monophosphate hydrolase activity(GO:0033699) |
| 1.6 | 4.9 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 1.6 | 14.6 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 1.6 | 41.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 1.6 | 11.1 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 1.5 | 6.2 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 1.5 | 4.5 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 1.5 | 7.5 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 1.4 | 15.3 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 1.4 | 38.8 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 1.4 | 10.9 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 1.3 | 9.4 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 1.3 | 4.0 | GO:0002046 | opsin binding(GO:0002046) |
| 1.3 | 6.6 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 1.3 | 7.9 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 1.3 | 20.8 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 1.2 | 9.9 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 1.2 | 9.9 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 1.2 | 21.4 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 1.2 | 4.8 | GO:0004335 | galactokinase activity(GO:0004335) |
| 1.2 | 3.5 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 1.2 | 12.8 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 1.2 | 9.2 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 1.1 | 3.4 | GO:0047315 | kynurenine-glyoxylate transaminase activity(GO:0047315) |
| 1.1 | 13.3 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 1.1 | 3.3 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 1.1 | 46.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 1.1 | 7.4 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 1.0 | 18.8 | GO:0030957 | Tat protein binding(GO:0030957) |
| 1.0 | 43.6 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 1.0 | 18.7 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 1.0 | 5.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 1.0 | 25.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 1.0 | 33.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 1.0 | 8.8 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 1.0 | 14.6 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.9 | 6.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.9 | 125.5 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.9 | 8.1 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.9 | 19.5 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.9 | 46.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.9 | 47.8 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.9 | 4.3 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.9 | 5.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.9 | 15.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.9 | 6.0 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.8 | 8.5 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.8 | 9.3 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.8 | 39.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.8 | 3.3 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.8 | 4.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.8 | 2.4 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.8 | 6.4 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.8 | 4.8 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.8 | 2.4 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.8 | 4.0 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.8 | 9.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.8 | 6.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.8 | 3.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.8 | 40.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.7 | 2.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.7 | 6.0 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.7 | 16.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.7 | 2.2 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.7 | 14.0 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.7 | 5.8 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.7 | 5.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.7 | 0.7 | GO:0003909 | DNA ligase activity(GO:0003909) |
| 0.7 | 4.9 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.7 | 21.9 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.7 | 14.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.7 | 8.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.6 | 32.0 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.6 | 12.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.6 | 3.2 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.6 | 23.2 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.6 | 2.4 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.6 | 3.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.6 | 8.8 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.6 | 28.8 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.6 | 16.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.6 | 19.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.6 | 4.0 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.6 | 9.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.6 | 31.6 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) ATP-dependent helicase activity(GO:0008026) RNA-dependent ATPase activity(GO:0008186) purine NTP-dependent helicase activity(GO:0070035) |
| 0.6 | 6.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.6 | 11.7 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.5 | 28.8 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.5 | 27.8 | GO:0016859 | cis-trans isomerase activity(GO:0016859) |
| 0.5 | 3.6 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.5 | 11.9 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.5 | 3.4 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.5 | 1.0 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.5 | 1.4 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.5 | 8.2 | GO:0004549 | tRNA-specific ribonuclease activity(GO:0004549) |
| 0.5 | 2.4 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.5 | 5.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.5 | 1.4 | GO:0004577 | N-acetylglucosaminyldiphosphodolichol N-acetylglucosaminyltransferase activity(GO:0004577) |
| 0.5 | 6.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.4 | 1.3 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.4 | 5.2 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.4 | 3.0 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.4 | 3.8 | GO:0030884 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.4 | 6.8 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.4 | 9.2 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.4 | 0.4 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.4 | 11.1 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.4 | 1.6 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.4 | 5.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.4 | 10.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.4 | 4.3 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.4 | 6.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.4 | 7.3 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.4 | 3.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.4 | 3.0 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.4 | 12.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.4 | 4.8 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.4 | 1.1 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.4 | 6.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.4 | 18.9 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.3 | 4.1 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.3 | 4.6 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.3 | 2.9 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.3 | 1.6 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.3 | 10.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.3 | 9.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.3 | 8.7 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.3 | 1.8 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.3 | 2.9 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.3 | 25.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.3 | 6.8 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.3 | 17.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.3 | 26.3 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.3 | 2.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.3 | 4.4 | GO:0015250 | water channel activity(GO:0015250) |
| 0.3 | 1.6 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.3 | 2.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.3 | 0.8 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.3 | 0.8 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.3 | 10.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.3 | 12.1 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.3 | 6.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.3 | 0.5 | GO:0000992 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.2 | 3.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.2 | 3.7 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.2 | 5.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 0.7 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.2 | 3.7 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.2 | 8.0 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.2 | 3.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.2 | 8.3 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.2 | 6.3 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.2 | 14.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.2 | 2.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.2 | 45.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.2 | 1.6 | GO:0000987 | core promoter proximal region sequence-specific DNA binding(GO:0000987) |
| 0.2 | 10.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 0.8 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.2 | 3.0 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.2 | 6.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.2 | 3.4 | GO:0016849 | phosphorus-oxygen lyase activity(GO:0016849) |
| 0.2 | 25.2 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.2 | 3.1 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.2 | 8.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.2 | 13.7 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.2 | 0.8 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.2 | 2.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 4.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.2 | 5.9 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.2 | 1.8 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.1 | 2.4 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 1.6 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 3.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 2.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 49.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 2.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 1.5 | GO:0004568 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.1 | 9.5 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.1 | 2.2 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.1 | 4.0 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 1.0 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 0.5 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.1 | 1.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 2.0 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.1 | 0.3 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 1.6 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 2.8 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 2.7 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.1 | 1.8 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 7.9 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 1.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 2.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 28.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 0.8 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 12.0 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.1 | 1.8 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 1.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 21.4 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.1 | 0.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 1.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.9 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 3.0 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.1 | 2.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 6.0 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 1.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.3 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.9 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.4 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 4.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.7 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 3.3 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 0.2 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 6.9 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 4.0 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 1.1 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 12.3 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.3 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.2 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.0 | 0.3 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.8 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 1.4 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 1.0 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.1 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 99.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 2.0 | 46.8 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 1.4 | 88.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 1.4 | 51.5 | PID ATM PATHWAY | ATM pathway |
| 1.2 | 73.9 | PID ATR PATHWAY | ATR signaling pathway |
| 1.0 | 98.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 1.0 | 47.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.9 | 31.8 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.9 | 26.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.8 | 8.0 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.7 | 45.1 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.7 | 33.9 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.7 | 18.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.6 | 17.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.6 | 15.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.6 | 56.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.6 | 15.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.6 | 122.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.6 | 17.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.5 | 16.9 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.5 | 11.3 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.5 | 16.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.5 | 25.4 | PID FOXO PATHWAY | FoxO family signaling |
| 0.4 | 9.3 | PID EPO PATHWAY | EPO signaling pathway |
| 0.4 | 10.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.3 | 5.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.3 | 10.0 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.3 | 6.3 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.3 | 3.9 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.3 | 5.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.3 | 16.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.3 | 10.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.3 | 16.8 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.3 | 11.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.3 | 20.8 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.3 | 10.0 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.3 | 7.0 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.3 | 4.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.3 | 9.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.3 | 8.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.2 | 11.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 3.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.2 | 11.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.2 | 1.4 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.2 | 5.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.2 | 6.8 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.2 | 7.7 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 3.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 8.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 11.4 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 2.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 3.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 5.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 3.6 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 3.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 20.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 2.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.8 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 0.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.7 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.6 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 1.2 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 9.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 151.0 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 4.6 | 68.6 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 4.5 | 67.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 4.3 | 99.9 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 4.3 | 170.3 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 2.7 | 32.9 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 2.6 | 60.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 2.5 | 227.6 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 2.5 | 17.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 2.3 | 43.2 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 2.2 | 20.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 2.2 | 165.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 2.2 | 17.5 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 2.2 | 41.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 2.1 | 31.9 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 2.0 | 41.8 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 1.8 | 22.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 1.7 | 225.0 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 1.7 | 51.6 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 1.7 | 144.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 1.6 | 42.7 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 1.5 | 30.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 1.4 | 19.9 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 1.4 | 40.1 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 1.3 | 27.6 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 1.2 | 14.9 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 1.2 | 8.3 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 1.2 | 23.8 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 1.2 | 56.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 1.1 | 35.7 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 1.1 | 13.0 | REACTOME CDT1 ASSOCIATION WITH THE CDC6 ORC ORIGIN COMPLEX | Genes involved in CDT1 association with the CDC6:ORC:origin complex |
| 1.0 | 9.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 1.0 | 14.5 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 1.0 | 86.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.9 | 33.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.9 | 88.1 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.9 | 19.6 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.9 | 15.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.9 | 11.2 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.9 | 6.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.8 | 27.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.8 | 14.8 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.8 | 27.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.8 | 22.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.8 | 11.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.7 | 20.1 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.7 | 12.6 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.7 | 16.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.6 | 6.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.5 | 10.3 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.5 | 17.2 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.5 | 41.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.5 | 10.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.4 | 9.6 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.4 | 21.3 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.4 | 8.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.4 | 16.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.4 | 12.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.4 | 7.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.4 | 9.5 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.4 | 4.9 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.3 | 4.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 2.9 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.3 | 10.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.3 | 6.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.3 | 5.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.3 | 4.8 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.3 | 5.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.3 | 9.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.3 | 4.7 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.3 | 13.9 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.3 | 9.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.3 | 21.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.2 | 6.5 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.2 | 17.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 5.3 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.2 | 5.1 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.2 | 3.8 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.2 | 4.7 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.2 | 3.9 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.2 | 6.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 4.1 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 4.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.2 | 14.0 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.2 | 4.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.2 | 6.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 4.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.2 | 3.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 1.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.2 | 2.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 5.6 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 1.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 0.1 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 5.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 4.1 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 9.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 1.7 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 2.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 3.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 1.6 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 2.2 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.1 | 2.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 0.8 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 1.0 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.1 | 3.3 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 2.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.5 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 1.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 1.1 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.8 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 1.8 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.2 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |