Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
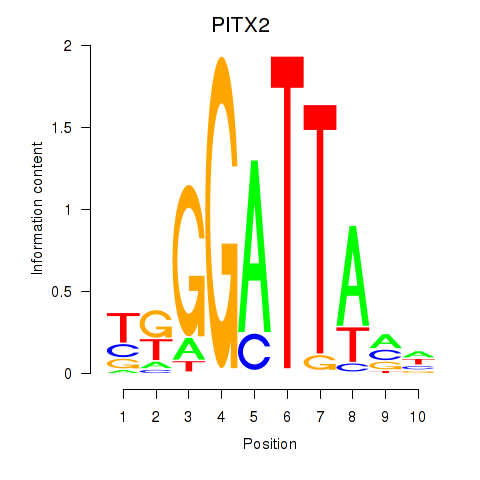
Results for PITX2
Z-value: 1.47
Transcription factors associated with PITX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PITX2
|
ENSG00000164093.17 | PITX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PITX2 | hg38_v1_chr4_-_110636963_110637042, hg38_v1_chr4_-_110641920_110642123, hg38_v1_chr4_-_110623051_110623085 | -0.11 | 1.2e-01 | Click! |
Activity profile of PITX2 motif
Sorted Z-values of PITX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PITX2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_35855627 | 10.13 |
ENST00000379893.5
|
DCLK1
|
doublecortin like kinase 1 |
| chr11_-_5227063 | 8.16 |
ENST00000335295.4
ENST00000485743.1 ENST00000647020.1 |
HBB
|
hemoglobin subunit beta |
| chr13_-_35855758 | 5.97 |
ENST00000615680.4
|
DCLK1
|
doublecortin like kinase 1 |
| chr11_-_133532493 | 5.14 |
ENST00000524381.6
|
OPCML
|
opioid binding protein/cell adhesion molecule like |
| chr4_-_101347492 | 4.90 |
ENST00000394854.8
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr4_-_101347327 | 4.52 |
ENST00000394853.8
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr3_+_159069252 | 4.14 |
ENST00000640015.1
ENST00000476809.7 ENST00000485419.7 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr4_-_101347471 | 3.98 |
ENST00000323055.10
ENST00000512215.5 |
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr4_-_101346842 | 3.98 |
ENST00000507176.5
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr12_-_89656051 | 3.51 |
ENST00000261173.6
|
ATP2B1
|
ATPase plasma membrane Ca2+ transporting 1 |
| chr12_-_89656093 | 3.42 |
ENST00000359142.7
|
ATP2B1
|
ATPase plasma membrane Ca2+ transporting 1 |
| chr16_-_11976611 | 2.49 |
ENST00000538896.5
ENST00000673243.1 |
NPIPB2
|
nuclear pore complex interacting protein family member B2 |
| chrX_-_6228835 | 2.31 |
ENST00000381095.8
|
NLGN4X
|
neuroligin 4 X-linked |
| chr12_+_69739370 | 1.96 |
ENST00000550536.5
ENST00000362025.9 |
RAB3IP
|
RAB3A interacting protein |
| chr11_+_61755372 | 1.72 |
ENST00000265460.9
|
MYRF
|
myelin regulatory factor |
| chr12_-_118359639 | 1.56 |
ENST00000541786.5
ENST00000419821.6 ENST00000541878.5 |
TAOK3
|
TAO kinase 3 |
| chr10_-_60141004 | 1.50 |
ENST00000355288.6
|
ANK3
|
ankyrin 3 |
| chr1_+_21977014 | 1.39 |
ENST00000337107.11
|
CELA3B
|
chymotrypsin like elastase 3B |
| chr14_+_75522531 | 1.37 |
ENST00000555504.1
|
BATF
|
basic leucine zipper ATF-like transcription factor |
| chr1_+_2476284 | 1.16 |
ENST00000378486.8
|
PLCH2
|
phospholipase C eta 2 |
| chr1_-_30757767 | 0.97 |
ENST00000294507.4
|
LAPTM5
|
lysosomal protein transmembrane 5 |
| chr1_+_2476315 | 0.91 |
ENST00000419816.6
|
PLCH2
|
phospholipase C eta 2 |
| chr21_-_18403754 | 0.86 |
ENST00000284885.8
|
TMPRSS15
|
transmembrane serine protease 15 |
| chr7_+_155458129 | 0.86 |
ENST00000297375.4
|
EN2
|
engrailed homeobox 2 |
| chr15_-_89221558 | 0.59 |
ENST00000268125.10
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr12_+_7789393 | 0.54 |
ENST00000229307.9
|
NANOG
|
Nanog homeobox |
| chr6_+_35342535 | 0.53 |
ENST00000360694.8
ENST00000418635.6 ENST00000448077.6 |
PPARD
|
peroxisome proliferator activated receptor delta |
| chr1_+_22001654 | 0.44 |
ENST00000290122.8
|
CELA3A
|
chymotrypsin like elastase 3A |
| chrX_+_15749848 | 0.43 |
ENST00000479740.5
ENST00000454127.2 |
CA5B
|
carbonic anhydrase 5B |
| chr6_-_53148822 | 0.42 |
ENST00000259803.8
|
GCM1
|
glial cells missing transcription factor 1 |
| chr19_+_2476118 | 0.33 |
ENST00000215631.9
ENST00000587345.1 |
GADD45B
|
growth arrest and DNA damage inducible beta |
| chr6_+_35342614 | 0.28 |
ENST00000337400.6
ENST00000311565.4 |
PPARD
|
peroxisome proliferator activated receptor delta |
| chr16_-_67980483 | 0.28 |
ENST00000268793.6
ENST00000672962.1 |
DPEP3
|
dipeptidase 3 |
| chr1_-_165445220 | 0.25 |
ENST00000619224.1
|
RXRG
|
retinoid X receptor gamma |
| chr9_-_14300231 | 0.25 |
ENST00000636735.1
|
NFIB
|
nuclear factor I B |
| chr7_-_128775793 | 0.24 |
ENST00000249389.3
|
OPN1SW
|
opsin 1, short wave sensitive |
| chr5_-_150758497 | 0.23 |
ENST00000521533.1
ENST00000424236.5 |
DCTN4
|
dynactin subunit 4 |
| chr3_+_154121366 | 0.20 |
ENST00000465093.6
ENST00000496710.5 ENST00000465817.1 |
ARHGEF26
|
Rho guanine nucleotide exchange factor 26 |
| chr6_+_31137646 | 0.20 |
ENST00000547221.1
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chr6_-_111483700 | 0.17 |
ENST00000435970.5
ENST00000358835.7 |
REV3L
|
REV3 like, DNA directed polymerase zeta catalytic subunit |
| chr1_-_165445088 | 0.12 |
ENST00000359842.10
|
RXRG
|
retinoid X receptor gamma |
| chr1_+_202010575 | 0.09 |
ENST00000367283.7
ENST00000367284.10 |
ELF3
|
E74 like ETS transcription factor 3 |
| chr9_+_34458752 | 0.08 |
ENST00000614641.4
ENST00000242317.9 ENST00000437363.5 |
DNAI1
|
dynein axonemal intermediate chain 1 |
| chr9_+_12693327 | 0.05 |
ENST00000388918.10
|
TYRP1
|
tyrosinase related protein 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 17.4 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 2.7 | 8.2 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 1.2 | 6.9 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.4 | 1.5 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.3 | 16.3 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.2 | 2.3 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.2 | 1.7 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.1 | 0.4 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.1 | 1.4 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 5.1 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.1 | 1.3 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.1 | 0.5 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.1 | 1.6 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 0.9 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 2.1 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.0 | 0.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.2 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) positive regulation of p38MAPK cascade(GO:1900745) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 8.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 1.6 | 17.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.3 | 6.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.5 | GO:0043194 | spectrin-associated cytoskeleton(GO:0014731) axon initial segment(GO:0043194) |
| 0.0 | 18.4 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 5.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.2 | GO:0042575 | DNA polymerase complex(GO:0042575) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 17.4 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 1.2 | 8.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.2 | 6.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 0.6 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 2.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.4 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 2.1 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 17.7 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 2.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.8 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.8 | GO:0030507 | spectrin binding(GO:0030507) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 17.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 5.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 17.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 8.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 1.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |