Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
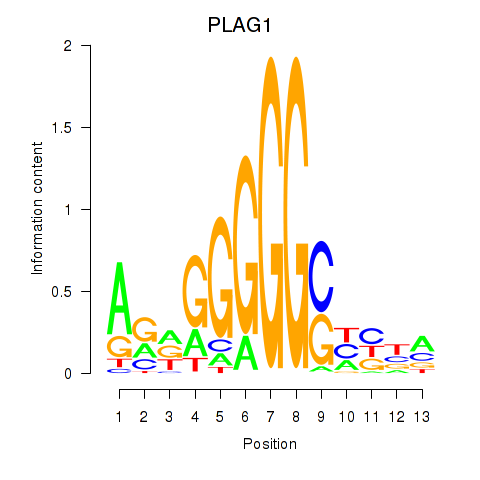
Results for PLAG1
Z-value: 3.56
Transcription factors associated with PLAG1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PLAG1
|
ENSG00000181690.8 | PLAG1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PLAG1 | hg38_v1_chr8_-_56211257_56211309 | 0.25 | 1.8e-04 | Click! |
Activity profile of PLAG1 motif
Sorted Z-values of PLAG1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PLAG1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_136980211 | 20.52 |
ENST00000444903.2
|
PTGDS
|
prostaglandin D2 synthase |
| chr6_-_83709019 | 19.47 |
ENST00000519779.5
ENST00000369694.6 ENST00000195649.10 |
SNAP91
|
synaptosome associated protein 91 |
| chr12_+_78864768 | 17.07 |
ENST00000261205.9
ENST00000457153.6 |
SYT1
|
synaptotagmin 1 |
| chr8_+_22054817 | 13.66 |
ENST00000432128.5
ENST00000443491.6 ENST00000517600.5 ENST00000523782.6 |
DMTN
|
dematin actin binding protein |
| chrX_+_111096211 | 11.41 |
ENST00000372010.5
ENST00000519681.5 |
PAK3
|
p21 (RAC1) activated kinase 3 |
| chr2_+_241188509 | 11.19 |
ENST00000674324.1
ENST00000274979.12 |
ANO7
|
anoctamin 7 |
| chr11_-_57514876 | 10.87 |
ENST00000528450.5
|
SLC43A1
|
solute carrier family 43 member 1 |
| chr5_+_161848112 | 10.61 |
ENST00000393943.10
|
GABRA1
|
gamma-aminobutyric acid type A receptor subunit alpha1 |
| chr11_-_73598067 | 10.41 |
ENST00000450446.6
ENST00000356467.5 |
FAM168A
|
family with sequence similarity 168 member A |
| chrX_+_111096136 | 10.34 |
ENST00000372007.10
|
PAK3
|
p21 (RAC1) activated kinase 3 |
| chrX_-_138711663 | 9.89 |
ENST00000315930.11
|
FGF13
|
fibroblast growth factor 13 |
| chr11_-_73598183 | 9.59 |
ENST00000064778.8
|
FAM168A
|
family with sequence similarity 168 member A |
| chr1_+_163068775 | 8.95 |
ENST00000421743.6
|
RGS4
|
regulator of G protein signaling 4 |
| chr5_+_161848536 | 8.79 |
ENST00000519621.2
ENST00000636573.1 |
GABRA1
|
gamma-aminobutyric acid type A receptor subunit alpha1 |
| chrX_+_52184874 | 8.59 |
ENST00000599522.7
ENST00000471932.6 |
MAGED4
|
MAGE family member D4 |
| chr5_+_161848314 | 8.40 |
ENST00000437025.6
|
GABRA1
|
gamma-aminobutyric acid type A receptor subunit alpha1 |
| chr22_+_29073112 | 8.16 |
ENST00000327813.9
ENST00000407188.5 |
KREMEN1
|
kringle containing transmembrane protein 1 |
| chr12_-_112382363 | 7.96 |
ENST00000682272.1
ENST00000377560.9 |
HECTD4
|
HECT domain E3 ubiquitin protein ligase 4 |
| chr7_-_158587710 | 7.82 |
ENST00000389416.8
|
PTPRN2
|
protein tyrosine phosphatase receptor type N2 |
| chr1_+_150257247 | 7.55 |
ENST00000647854.1
|
CA14
|
carbonic anhydrase 14 |
| chr19_+_4639505 | 7.22 |
ENST00000327473.9
|
TNFAIP8L1
|
TNF alpha induced protein 8 like 1 |
| chr5_-_55534955 | 7.19 |
ENST00000307259.9
ENST00000264775.9 |
PLPP1
|
phospholipid phosphatase 1 |
| chr16_-_29899532 | 7.00 |
ENST00000308713.9
ENST00000617533.5 |
SEZ6L2
|
seizure related 6 homolog like 2 |
| chr12_-_12266769 | 6.83 |
ENST00000543091.1
|
LRP6
|
LDL receptor related protein 6 |
| chr14_+_79279339 | 6.72 |
ENST00000557594.5
|
NRXN3
|
neurexin 3 |
| chr14_-_53153281 | 6.51 |
ENST00000357758.3
ENST00000673822.2 |
DDHD1
|
DDHD domain containing 1 |
| chr12_-_27780236 | 6.37 |
ENST00000381273.4
|
MANSC4
|
MANSC domain containing 4 |
| chr14_+_79279906 | 6.29 |
ENST00000428277.6
|
NRXN3
|
neurexin 3 |
| chr12_-_52321395 | 6.15 |
ENST00000293670.3
|
KRT83
|
keratin 83 |
| chr14_-_81220951 | 6.13 |
ENST00000553612.6
|
GTF2A1
|
general transcription factor IIA subunit 1 |
| chr2_-_152098810 | 5.94 |
ENST00000636442.1
ENST00000638005.1 |
CACNB4
|
calcium voltage-gated channel auxiliary subunit beta 4 |
| chrX_-_45200828 | 5.91 |
ENST00000398000.7
|
DIPK2B
|
divergent protein kinase domain 2B |
| chr14_-_53153098 | 5.78 |
ENST00000612692.4
ENST00000395606.5 |
DDHD1
|
DDHD domain containing 1 |
| chr9_-_132944600 | 5.76 |
ENST00000490179.3
ENST00000643583.1 ENST00000298552.9 ENST00000643072.1 ENST00000642745.1 ENST00000647462.1 ENST00000643875.1 ENST00000642627.1 ENST00000475903.6 ENST00000642617.1 ENST00000642646.1 ENST00000646625.1 ENST00000645150.1 ENST00000645129.1 ENST00000403810.6 ENST00000643691.1 ENST00000644097.1 |
TSC1
|
TSC complex subunit 1 |
| chr2_-_152098670 | 5.70 |
ENST00000636129.1
ENST00000636785.1 ENST00000636496.1 |
CACNB4
|
calcium voltage-gated channel auxiliary subunit beta 4 |
| chr7_+_73328177 | 5.58 |
ENST00000442793.5
ENST00000413573.6 |
FKBP6
|
FKBP prolyl isomerase family member 6 (inactive) |
| chr3_-_51967410 | 5.45 |
ENST00000461554.6
ENST00000483411.5 ENST00000461544.2 ENST00000355852.6 |
PCBP4
|
poly(rC) binding protein 4 |
| chr17_-_4704040 | 5.42 |
ENST00000570571.5
ENST00000575101.1 ENST00000574876.5 ENST00000572293.7 |
PELP1
|
proline, glutamate and leucine rich protein 1 |
| chr16_-_29899043 | 5.37 |
ENST00000346932.9
ENST00000350527.7 ENST00000568380.1 |
SEZ6L2
|
seizure related 6 homolog like 2 |
| chr6_-_29628038 | 5.37 |
ENST00000355973.7
ENST00000377012.8 |
GABBR1
|
gamma-aminobutyric acid type B receptor subunit 1 |
| chr10_-_73811583 | 5.36 |
ENST00000309979.11
|
NDST2
|
N-deacetylase and N-sulfotransferase 2 |
| chr3_-_51968387 | 5.31 |
ENST00000490063.5
ENST00000468324.5 ENST00000497653.5 ENST00000484633.5 |
PCBP4
|
poly(rC) binding protein 4 |
| chr14_+_79279403 | 5.23 |
ENST00000281127.11
|
NRXN3
|
neurexin 3 |
| chr16_-_29899245 | 5.16 |
ENST00000537485.5
|
SEZ6L2
|
seizure related 6 homolog like 2 |
| chr19_-_38229654 | 5.13 |
ENST00000412732.5
ENST00000456296.5 |
DPF1
|
double PHD fingers 1 |
| chr19_-_13906062 | 5.01 |
ENST00000591586.5
ENST00000346736.6 |
BRME1
|
break repair meiotic recombinase recruitment factor 1 |
| chr19_+_40467145 | 5.00 |
ENST00000338932.7
|
SPTBN4
|
spectrin beta, non-erythrocytic 4 |
| chr7_+_73328152 | 4.99 |
ENST00000252037.5
ENST00000431982.6 |
FKBP6
|
FKBP prolyl isomerase family member 6 (inactive) |
| chr11_+_46277648 | 4.99 |
ENST00000621158.5
|
CREB3L1
|
cAMP responsive element binding protein 3 like 1 |
| chr17_-_76141240 | 4.91 |
ENST00000322957.7
|
FOXJ1
|
forkhead box J1 |
| chr14_-_21526312 | 4.80 |
ENST00000537235.2
|
SALL2
|
spalt like transcription factor 2 |
| chr6_-_32224060 | 4.66 |
ENST00000375023.3
|
NOTCH4
|
notch receptor 4 |
| chr2_-_152099023 | 4.64 |
ENST00000201943.10
ENST00000427385.6 ENST00000539935.7 |
CACNB4
|
calcium voltage-gated channel auxiliary subunit beta 4 |
| chr14_+_79279681 | 4.63 |
ENST00000679122.1
|
NRXN3
|
neurexin 3 |
| chr2_-_74391837 | 4.50 |
ENST00000417090.1
ENST00000409868.5 ENST00000680606.1 |
DCTN1
|
dynactin subunit 1 |
| chr19_+_40466976 | 4.48 |
ENST00000598249.6
|
SPTBN4
|
spectrin beta, non-erythrocytic 4 |
| chr9_-_120714457 | 4.47 |
ENST00000373930.4
|
MEGF9
|
multiple EGF like domains 9 |
| chrX_+_52184904 | 4.40 |
ENST00000375626.7
ENST00000467526.1 |
MAGED4
|
MAGE family member D4 |
| chr1_-_31704001 | 4.39 |
ENST00000373672.8
|
COL16A1
|
collagen type XVI alpha 1 chain |
| chr14_+_79280056 | 4.35 |
ENST00000676811.1
|
NRXN3
|
neurexin 3 |
| chr1_-_115338231 | 4.32 |
ENST00000369512.3
ENST00000680116.1 ENST00000681124.1 ENST00000675637.2 ENST00000676038.2 |
NGF
|
nerve growth factor |
| chr5_-_59893718 | 4.28 |
ENST00000340635.11
|
PDE4D
|
phosphodiesterase 4D |
| chr2_-_152099155 | 4.27 |
ENST00000637309.1
|
CACNB4
|
calcium voltage-gated channel auxiliary subunit beta 4 |
| chr19_-_461007 | 4.22 |
ENST00000264554.11
|
SHC2
|
SHC adaptor protein 2 |
| chr17_+_75109939 | 4.19 |
ENST00000581078.1
ENST00000245543.6 ENST00000582136.5 |
ARMC7
|
armadillo repeat containing 7 |
| chr2_-_74392025 | 4.15 |
ENST00000440727.1
ENST00000409240.5 |
DCTN1
|
dynactin subunit 1 |
| chr10_-_97687191 | 4.11 |
ENST00000370626.4
|
AVPI1
|
arginine vasopressin induced 1 |
| chr20_-_18057841 | 4.11 |
ENST00000278780.7
|
OVOL2
|
ovo like zinc finger 2 |
| chr16_+_31117656 | 4.10 |
ENST00000219797.9
ENST00000448516.6 |
KAT8
|
lysine acetyltransferase 8 |
| chr12_-_57738740 | 4.06 |
ENST00000547588.6
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr21_-_36966612 | 3.75 |
ENST00000674895.3
|
HLCS
|
holocarboxylase synthetase |
| chr17_+_49969178 | 3.68 |
ENST00000240306.5
|
DLX4
|
distal-less homeobox 4 |
| chr19_-_38229714 | 3.54 |
ENST00000416611.5
|
DPF1
|
double PHD fingers 1 |
| chr10_+_110007964 | 3.50 |
ENST00000277900.12
ENST00000356080.9 |
ADD3
|
adducin 3 |
| chr6_-_31972290 | 3.36 |
ENST00000375349.7
|
DXO
|
decapping exoribonuclease |
| chr12_-_48004496 | 3.31 |
ENST00000337299.7
|
COL2A1
|
collagen type II alpha 1 chain |
| chr14_-_74612226 | 3.25 |
ENST00000261978.9
|
LTBP2
|
latent transforming growth factor beta binding protein 2 |
| chr2_-_73284431 | 3.24 |
ENST00000521871.5
ENST00000520530.3 |
FBXO41
|
F-box protein 41 |
| chr22_+_29205877 | 3.21 |
ENST00000334018.11
ENST00000429226.5 ENST00000404755.7 ENST00000404820.7 ENST00000430127.1 |
EMID1
|
EMI domain containing 1 |
| chr4_+_54229261 | 3.14 |
ENST00000508170.5
ENST00000512143.1 ENST00000257290.10 |
PDGFRA
|
platelet derived growth factor receptor alpha |
| chr1_-_204685700 | 3.14 |
ENST00000367177.4
|
LRRN2
|
leucine rich repeat neuronal 2 |
| chr9_+_129081098 | 3.10 |
ENST00000406974.7
ENST00000372546.9 |
DOLPP1
|
dolichyldiphosphatase 1 |
| chr13_+_53028806 | 3.06 |
ENST00000219022.3
|
OLFM4
|
olfactomedin 4 |
| chrX_-_72307148 | 3.06 |
ENST00000453707.6
ENST00000373619.7 |
CITED1
|
Cbp/p300 interacting transactivator with Glu/Asp rich carboxy-terminal domain 1 |
| chr4_+_61200318 | 3.04 |
ENST00000683033.1
|
ADGRL3
|
adhesion G protein-coupled receptor L3 |
| chr16_+_425599 | 2.96 |
ENST00000262305.9
|
RAB11FIP3
|
RAB11 family interacting protein 3 |
| chr12_-_12267003 | 2.91 |
ENST00000535731.1
ENST00000261349.9 |
LRP6
|
LDL receptor related protein 6 |
| chr4_+_41990496 | 2.88 |
ENST00000264451.12
|
SLC30A9
|
solute carrier family 30 member 9 |
| chr12_-_48004467 | 2.87 |
ENST00000380518.8
|
COL2A1
|
collagen type II alpha 1 chain |
| chr19_-_4535221 | 2.85 |
ENST00000381848.7
ENST00000586133.1 |
PLIN5
|
perilipin 5 |
| chr2_-_29921580 | 2.78 |
ENST00000389048.8
|
ALK
|
ALK receptor tyrosine kinase |
| chr1_-_110070654 | 2.71 |
ENST00000647563.2
|
ALX3
|
ALX homeobox 3 |
| chr9_+_69325168 | 2.70 |
ENST00000303068.14
|
FAM189A2
|
family with sequence similarity 189 member A2 |
| chr7_-_27152561 | 2.70 |
ENST00000467897.2
ENST00000612286.5 ENST00000498652.1 |
ENSG00000273433.1
HOXA3
ENSG00000272801.1
|
novel transcript homeobox A3 novel transcript |
| chr1_-_243850216 | 2.67 |
ENST00000673466.1
|
AKT3
|
AKT serine/threonine kinase 3 |
| chr9_-_133129395 | 2.65 |
ENST00000393157.8
|
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chr6_-_31972123 | 2.60 |
ENST00000337523.10
|
DXO
|
decapping exoribonuclease |
| chr7_-_100573865 | 2.55 |
ENST00000622764.3
|
SAP25
|
Sin3A associated protein 25 |
| chr14_-_25050111 | 2.46 |
ENST00000323944.9
|
STXBP6
|
syntaxin binding protein 6 |
| chr2_-_178072751 | 2.45 |
ENST00000286063.11
|
PDE11A
|
phosphodiesterase 11A |
| chr12_-_108339300 | 2.29 |
ENST00000550402.6
ENST00000552995.5 ENST00000312143.11 |
CMKLR1
|
chemerin chemokine-like receptor 1 |
| chr2_+_240625237 | 2.20 |
ENST00000407714.1
|
GPR35
|
G protein-coupled receptor 35 |
| chr3_-_73624840 | 2.15 |
ENST00000308537.4
ENST00000263666.9 |
PDZRN3
|
PDZ domain containing ring finger 3 |
| chr14_-_25049889 | 2.05 |
ENST00000419632.6
ENST00000396700.5 |
STXBP6
|
syntaxin binding protein 6 |
| chr3_-_15332526 | 2.04 |
ENST00000383791.8
|
SH3BP5
|
SH3 domain binding protein 5 |
| chr19_+_3359563 | 1.99 |
ENST00000589123.5
ENST00000395111.7 ENST00000586919.5 |
NFIC
|
nuclear factor I C |
| chr6_-_36986122 | 1.90 |
ENST00000460219.2
ENST00000373627.10 ENST00000373616.9 |
MTCH1
|
mitochondrial carrier 1 |
| chr19_+_38433676 | 1.89 |
ENST00000359596.8
ENST00000355481.8 |
RYR1
|
ryanodine receptor 1 |
| chr16_+_33827140 | 1.89 |
ENST00000562905.2
|
IGHV3OR16-13
|
immunoglobulin heavy variable 3/OR16-13 (non-functional) |
| chr16_+_285680 | 1.89 |
ENST00000435833.1
|
PDIA2
|
protein disulfide isomerase family A member 2 |
| chr1_+_4654732 | 1.88 |
ENST00000378190.7
|
AJAP1
|
adherens junctions associated protein 1 |
| chr11_+_2301987 | 1.88 |
ENST00000612299.4
ENST00000182290.9 |
TSPAN32
|
tetraspanin 32 |
| chr3_+_135965718 | 1.77 |
ENST00000264977.8
ENST00000490467.5 |
PPP2R3A
|
protein phosphatase 2 regulatory subunit B''alpha |
| chr1_-_160070102 | 1.75 |
ENST00000638728.1
ENST00000637644.1 |
KCNJ10
|
potassium inwardly rectifying channel subfamily J member 10 |
| chr1_-_160070150 | 1.72 |
ENST00000644903.1
|
KCNJ10
|
potassium inwardly rectifying channel subfamily J member 10 |
| chr9_-_137032081 | 1.71 |
ENST00000314412.7
|
FUT7
|
fucosyltransferase 7 |
| chr14_-_31026363 | 1.66 |
ENST00000357479.10
ENST00000355683.9 |
STRN3
|
striatin 3 |
| chr1_+_2476284 | 1.60 |
ENST00000378486.8
|
PLCH2
|
phospholipase C eta 2 |
| chr11_+_71527267 | 1.59 |
ENST00000398536.6
|
KRTAP5-7
|
keratin associated protein 5-7 |
| chr1_+_2476315 | 1.57 |
ENST00000419816.6
|
PLCH2
|
phospholipase C eta 2 |
| chr10_-_70283998 | 1.52 |
ENST00000277942.7
|
NPFFR1
|
neuropeptide FF receptor 1 |
| chr16_+_14833713 | 1.42 |
ENST00000287667.12
ENST00000620755.4 ENST00000610363.4 |
NOMO1
|
NODAL modulator 1 |
| chrX_+_150361559 | 1.39 |
ENST00000262858.8
|
MAMLD1
|
mastermind like domain containing 1 |
| chr6_+_106086316 | 1.36 |
ENST00000369091.6
ENST00000369096.9 |
PRDM1
|
PR/SET domain 1 |
| chr2_+_156436423 | 1.36 |
ENST00000540309.5
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 |
| chr19_+_13906190 | 1.26 |
ENST00000318003.11
|
CC2D1A
|
coiled-coil and C2 domain containing 1A |
| chr20_+_58891981 | 1.22 |
ENST00000488652.6
ENST00000476935.6 ENST00000492907.6 ENST00000603546.2 |
GNAS
|
GNAS complex locus |
| chr3_-_116444983 | 1.17 |
ENST00000333617.8
|
LSAMP
|
limbic system associated membrane protein |
| chr19_+_13906255 | 1.17 |
ENST00000589606.5
|
CC2D1A
|
coiled-coil and C2 domain containing 1A |
| chr20_+_1895365 | 1.15 |
ENST00000358771.5
|
SIRPA
|
signal regulatory protein alpha |
| chr20_+_36573458 | 1.08 |
ENST00000373874.6
|
TGIF2
|
TGFB induced factor homeobox 2 |
| chr11_+_118607579 | 1.04 |
ENST00000530708.4
|
PHLDB1
|
pleckstrin homology like domain family B member 1 |
| chr6_-_35497042 | 1.03 |
ENST00000639578.3
ENST00000338863.13 |
TEAD3
|
TEA domain transcription factor 3 |
| chr19_-_43754901 | 0.98 |
ENST00000270066.11
ENST00000601170.5 |
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr1_-_24143112 | 0.92 |
ENST00000270800.2
|
IL22RA1
|
interleukin 22 receptor subunit alpha 1 |
| chr6_-_70303070 | 0.91 |
ENST00000370496.3
ENST00000357250.11 |
COL9A1
|
collagen type IX alpha 1 chain |
| chr1_-_19980416 | 0.88 |
ENST00000375111.7
|
PLA2G2A
|
phospholipase A2 group IIA |
| chr19_+_55385928 | 0.79 |
ENST00000431533.6
ENST00000428193.6 ENST00000558815.5 ENST00000344063.7 ENST00000560583.5 ENST00000560055.5 ENST00000559463.5 |
RPL28
|
ribosomal protein L28 |
| chr1_-_92483947 | 0.76 |
ENST00000370332.5
|
GFI1
|
growth factor independent 1 transcriptional repressor |
| chr17_+_66835110 | 0.75 |
ENST00000533854.6
|
CACNG5
|
calcium voltage-gated channel auxiliary subunit gamma 5 |
| chr16_+_56451513 | 0.71 |
ENST00000562150.5
ENST00000561646.5 ENST00000566157.6 ENST00000568397.1 |
OGFOD1
|
2-oxoglutarate and iron dependent oxygenase domain containing 1 |
| chr17_+_4951080 | 0.69 |
ENST00000521811.5
ENST00000323997.10 ENST00000522249.5 ENST00000519584.5 ENST00000519602.6 |
ENO3
|
enolase 3 |
| chr17_-_28812224 | 0.68 |
ENST00000582059.6
|
FAM222B
|
family with sequence similarity 222 member B |
| chr7_-_149773207 | 0.64 |
ENST00000484747.5
|
ZNF467
|
zinc finger protein 467 |
| chr17_-_1400168 | 0.63 |
ENST00000573026.1
ENST00000575977.1 ENST00000571732.5 ENST00000264335.13 |
YWHAE
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein epsilon |
| chr20_-_22584547 | 0.59 |
ENST00000419308.7
|
FOXA2
|
forkhead box A2 |
| chr11_-_64744317 | 0.59 |
ENST00000419843.1
ENST00000394430.5 |
RASGRP2
|
RAS guanyl releasing protein 2 |
| chr5_-_177509814 | 0.58 |
ENST00000510898.7
ENST00000502885.5 ENST00000506493.5 |
DOK3
|
docking protein 3 |
| chr3_-_116445458 | 0.58 |
ENST00000490035.7
|
LSAMP
|
limbic system associated membrane protein |
| chr6_+_31971831 | 0.51 |
ENST00000375331.7
ENST00000375333.3 |
STK19
|
serine/threonine kinase 19 |
| chr10_+_134465 | 0.50 |
ENST00000439456.5
ENST00000397962.8 ENST00000309776.8 ENST00000397959.7 |
ZMYND11
|
zinc finger MYND-type containing 11 |
| chr11_+_64285839 | 0.50 |
ENST00000438980.7
ENST00000540370.1 |
GPR137
|
G protein-coupled receptor 137 |
| chr14_-_21526391 | 0.49 |
ENST00000611430.4
|
SALL2
|
spalt like transcription factor 2 |
| chr2_-_109614143 | 0.48 |
ENST00000356688.8
|
SEPTIN10
|
septin 10 |
| chr1_-_44017296 | 0.39 |
ENST00000357730.6
ENST00000360584.6 ENST00000528803.1 |
SLC6A9
|
solute carrier family 6 member 9 |
| chr5_+_175658008 | 0.39 |
ENST00000377291.2
|
HRH2
|
histamine receptor H2 |
| chr3_+_19947074 | 0.33 |
ENST00000273047.9
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr11_+_57338344 | 0.28 |
ENST00000263314.3
ENST00000616487.4 |
P2RX3
|
purinergic receptor P2X 3 |
| chr1_+_202462730 | 0.20 |
ENST00000290419.9
ENST00000491336.5 |
PPP1R12B
|
protein phosphatase 1 regulatory subunit 12B |
| chr2_-_127642131 | 0.13 |
ENST00000426981.5
|
LIMS2
|
LIM zinc finger domain containing 2 |
| chr4_-_98929092 | 0.12 |
ENST00000280892.10
ENST00000511644.5 ENST00000504432.5 ENST00000450253.7 ENST00000505992.1 |
EIF4E
|
eukaryotic translation initiation factor 4E |
| chr19_+_42284483 | 0.12 |
ENST00000575354.6
|
CIC
|
capicua transcriptional repressor |
| chr17_-_28812392 | 0.07 |
ENST00000452648.8
|
FAM222B
|
family with sequence similarity 222 member B |
| chr7_-_149773548 | 0.03 |
ENST00000302017.4
|
ZNF467
|
zinc finger protein 467 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.7 | 20.0 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 5.7 | 17.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 3.7 | 11.2 | GO:0061590 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 3.2 | 9.7 | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 2.3 | 9.3 | GO:1990834 | response to odorant(GO:1990834) |
| 2.0 | 13.7 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 1.5 | 10.6 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 1.2 | 5.8 | GO:0051029 | rRNA transport(GO:0051029) |
| 1.1 | 8.7 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 1.0 | 3.1 | GO:0072275 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 1.0 | 27.8 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 1.0 | 4.9 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.9 | 9.5 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.9 | 21.7 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.9 | 3.8 | GO:0009305 | protein biotinylation(GO:0009305) histone biotinylation(GO:0071110) |
| 0.9 | 18.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.9 | 27.2 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.8 | 4.1 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.7 | 7.2 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.7 | 2.8 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.7 | 2.8 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.7 | 3.5 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.7 | 2.7 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.6 | 19.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.6 | 5.0 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.5 | 4.3 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.5 | 4.7 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.5 | 3.1 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.5 | 4.3 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.5 | 1.9 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.5 | 1.4 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 0.5 | 4.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.4 | 3.1 | GO:0046465 | dolichyl diphosphate biosynthetic process(GO:0006489) dolichyl diphosphate metabolic process(GO:0046465) |
| 0.4 | 5.4 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.4 | 12.0 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.4 | 3.0 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.4 | 6.2 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.3 | 1.7 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.3 | 6.0 | GO:0071028 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.3 | 22.8 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.3 | 1.9 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.3 | 1.4 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.3 | 3.1 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.3 | 0.8 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.2 | 11.3 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.2 | 3.0 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.2 | 0.6 | GO:0003064 | regulation of heart rate by hormone(GO:0003064) negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.2 | 1.9 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.2 | 1.0 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.2 | 4.1 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.2 | 0.6 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.2 | 5.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 | 4.4 | GO:0033622 | integrin activation(GO:0033622) |
| 0.2 | 1.9 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.2 | 7.2 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.2 | 2.7 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 8.9 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.1 | 1.9 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.1 | 7.8 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 7.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 2.3 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.1 | 2.4 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 10.8 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.1 | 1.1 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 1.7 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 | 1.0 | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 | 5.4 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 1.8 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.1 | 7.6 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.1 | 1.2 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 2.7 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.1 | 0.3 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 0.1 | 6.1 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 6.2 | GO:0042633 | molting cycle(GO:0042303) hair cycle(GO:0042633) |
| 0.1 | 3.3 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 0.3 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.1 | 0.9 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.1 | 0.7 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.1 | 2.0 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 0.7 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 8.3 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 2.1 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 3.2 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.0 | 1.8 | GO:0061053 | somite development(GO:0061053) |
| 0.0 | 2.0 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 4.1 | GO:0042177 | negative regulation of protein catabolic process(GO:0042177) |
| 0.0 | 8.0 | GO:0033500 | carbohydrate homeostasis(GO:0033500) glucose homeostasis(GO:0042593) |
| 0.0 | 0.7 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.0 | 0.4 | GO:0001696 | gastric acid secretion(GO:0001696) |
| 0.0 | 1.5 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.5 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 0.9 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 17.1 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 2.4 | 9.7 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 1.4 | 5.8 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 1.3 | 5.4 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.9 | 9.5 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.7 | 27.8 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.7 | 6.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.7 | 5.3 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.6 | 8.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.5 | 13.7 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) platelet dense tubular network membrane(GO:0031095) |
| 0.5 | 4.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.5 | 6.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.4 | 24.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.4 | 1.9 | GO:0030314 | junctional membrane complex(GO:0030314) ryanodine receptor complex(GO:1990425) |
| 0.3 | 1.9 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.3 | 4.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 8.7 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.2 | 10.6 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.2 | 1.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 4.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.2 | 5.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.2 | 3.8 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.2 | 19.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.2 | 19.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 1.0 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 8.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 31.1 | GO:0098793 | presynapse(GO:0098793) |
| 0.1 | 0.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 3.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 2.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 2.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 8.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 3.5 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 3.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 3.0 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.9 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 3.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 3.1 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 3.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 2.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.7 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 1.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 2.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 71.1 | GO:0031224 | intrinsic component of membrane(GO:0031224) |
| 0.0 | 0.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.6 | GO:0005871 | kinesin complex(GO:0005871) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 20.5 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 3.4 | 17.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 3.2 | 9.7 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 3.1 | 27.8 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 1.5 | 6.0 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 1.3 | 5.4 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 1.3 | 5.4 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 1.2 | 7.2 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 1.1 | 4.3 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) |
| 1.0 | 3.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 1.0 | 27.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.9 | 3.8 | GO:0018271 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 0.9 | 6.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.8 | 19.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.7 | 11.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.7 | 2.8 | GO:0035473 | lipase binding(GO:0035473) |
| 0.6 | 20.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.6 | 21.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.5 | 4.1 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.5 | 1.9 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.5 | 2.8 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.4 | 7.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.4 | 23.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.4 | 10.6 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.4 | 3.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.3 | 1.4 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.3 | 8.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.3 | 11.3 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.2 | 5.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 7.8 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 0.9 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.2 | 5.8 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.2 | 4.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 4.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 0.6 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.2 | 2.4 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.2 | 3.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.2 | 1.7 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.2 | 5.5 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.2 | 3.0 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.2 | 2.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.2 | 9.9 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 1.9 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.7 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.1 | 16.3 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.1 | 0.9 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 3.9 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 6.1 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 1.7 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 2.9 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 10.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 0.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 2.3 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 4.2 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 1.5 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 10.1 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.7 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 1.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.6 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 3.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 3.3 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 0.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 17.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.4 | 21.7 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.3 | 17.1 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.2 | 11.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 7.0 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.2 | 8.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 3.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 4.2 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 4.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 5.4 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 3.0 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 4.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 0.9 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.9 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 8.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 2.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 2.0 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 2.6 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 1.2 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 6.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.9 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 2.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 17.1 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.7 | 27.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.5 | 4.3 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.5 | 7.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.4 | 3.1 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.4 | 4.7 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.4 | 27.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.3 | 5.8 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.3 | 5.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 2.7 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 9.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 10.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 10.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.2 | 4.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 3.1 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.2 | 3.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 5.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 10.2 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 8.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 6.1 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 4.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 2.6 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 1.4 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 4.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 0.9 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 2.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.6 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 1.1 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |