Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for PRDM1
Z-value: 0.35
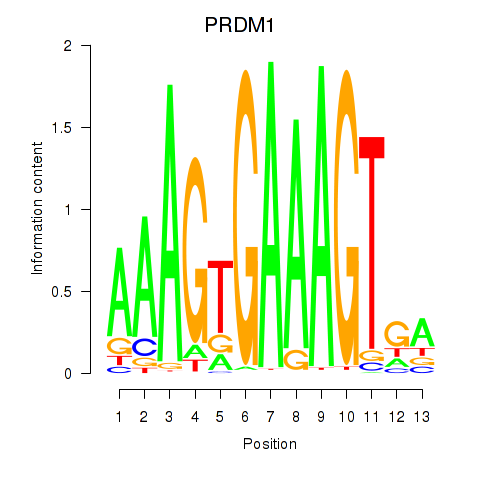
Transcription factors associated with PRDM1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PRDM1
|
ENSG00000057657.17 | PRDM1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PRDM1 | hg38_v1_chr6_+_106087580_106087762, hg38_v1_chr6_+_106086316_106086349, hg38_v1_chr6_+_106098933_106098958 | 0.19 | 4.4e-03 | Click! |
Activity profile of PRDM1 motif
Sorted Z-values of PRDM1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PRDM1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 16.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 3.1 | 3.1 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 2.3 | 16.0 | GO:0030421 | defecation(GO:0030421) |
| 2.2 | 6.6 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 2.1 | 6.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 1.6 | 8.2 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 1.6 | 4.9 | GO:1904582 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 1.6 | 17.3 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 1.5 | 10.6 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 1.3 | 5.4 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 1.2 | 4.6 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 1.2 | 4.6 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 1.0 | 3.1 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 1.0 | 3.0 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.9 | 3.7 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.9 | 2.8 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.9 | 5.4 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.8 | 7.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.7 | 3.5 | GO:2000542 | negative regulation of gastrulation(GO:2000542) |
| 0.7 | 5.6 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.7 | 2.0 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.7 | 4.7 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.6 | 1.9 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.6 | 2.5 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.5 | 4.3 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.5 | 2.0 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.5 | 1.5 | GO:1901877 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.5 | 1.4 | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) |
| 0.4 | 1.9 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.4 | 3.1 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.4 | 1.5 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.4 | 2.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.4 | 1.5 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.4 | 2.9 | GO:0015747 | urate transport(GO:0015747) |
| 0.4 | 1.1 | GO:0002876 | positive regulation of chronic inflammatory response to antigenic stimulus(GO:0002876) |
| 0.4 | 1.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.3 | 0.7 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.3 | 4.1 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.3 | 33.9 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.3 | 1.6 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.3 | 2.5 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.3 | 3.6 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.3 | 0.8 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.3 | 1.8 | GO:1901098 | regulation of autophagosome maturation(GO:1901096) positive regulation of autophagosome maturation(GO:1901098) |
| 0.3 | 2.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.3 | 7.3 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.2 | 1.8 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.2 | 0.9 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.2 | 4.9 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.2 | 0.6 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.2 | 2.4 | GO:0048625 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) myoblast fate commitment(GO:0048625) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.2 | 1.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.2 | 2.5 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.2 | 0.6 | GO:0044007 | negative regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010768) transmission of virus(GO:0019089) dissemination or transmission of symbiont from host(GO:0044007) dissemination or transmission of organism from other organism involved in symbiotic interaction(GO:0051821) |
| 0.2 | 2.0 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.2 | 2.2 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.1 | 2.9 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 0.6 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 2.1 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 2.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 0.8 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.1 | 2.0 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 1.6 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.1 | 1.2 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.3 | GO:0002678 | positive regulation of chronic inflammatory response(GO:0002678) |
| 0.1 | 1.5 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 0.9 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.8 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 1.1 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 2.2 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 1.8 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.0 | 2.1 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 4.0 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 1.8 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.9 | GO:0001696 | gastric acid secretion(GO:0001696) |
| 0.0 | 0.3 | GO:0045345 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.0 | 0.9 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.9 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.1 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.0 | 0.4 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.2 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 1.7 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 4.0 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.0 | 1.1 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.4 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 28.0 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 2.5 | 10.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 2.5 | 7.4 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 2.2 | 6.6 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 1.8 | 8.8 | GO:0042825 | TAP complex(GO:0042825) |
| 1.7 | 6.9 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 1.2 | 3.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.7 | 8.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.7 | 2.0 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.5 | 1.5 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.3 | 2.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.3 | 16.1 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.3 | 2.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.3 | 2.9 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 2.4 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.2 | 1.8 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.2 | 1.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.2 | 1.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 6.9 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 7.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 1.5 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 1.2 | GO:0000938 | GARP complex(GO:0000938) |
| 0.1 | 3.7 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 0.3 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.6 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 1.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.9 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 2.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.5 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 0.8 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 1.5 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 0.2 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.1 | 4.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.0 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 1.8 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 2.7 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 1.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 1.1 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.7 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 1.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 13.3 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 3.2 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 2.7 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 8.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 17.9 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 2.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 1.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 19.0 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.8 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 1.5 | 7.4 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 1.4 | 10.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 1.2 | 16.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 1.2 | 4.9 | GO:0005151 | interleukin-1, Type II receptor binding(GO:0005151) |
| 1.1 | 5.3 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.9 | 7.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.9 | 5.6 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.9 | 5.4 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.8 | 23.8 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.7 | 3.6 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.7 | 2.8 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.7 | 8.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.6 | 4.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.5 | 4.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.5 | 1.5 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.4 | 6.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.4 | 1.5 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.3 | 1.4 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.3 | 2.9 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.3 | 1.6 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.3 | 1.8 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.3 | 3.5 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.3 | 4.9 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.3 | 9.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 2.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.2 | 12.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 1.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.2 | 6.2 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.2 | 2.0 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 0.6 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.2 | 1.9 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.2 | 3.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 3.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 4.5 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 4.7 | GO:0001972 | retinoic acid binding(GO:0001972) glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 2.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.8 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 1.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.9 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 3.7 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.8 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 0.9 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 1.5 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.1 | 2.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.3 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.1 | 2.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 0.6 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 4.7 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 3.0 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 16.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 2.0 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 1.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 6.2 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.1 | 0.6 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 2.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 7.6 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 8.8 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 2.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.7 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 0.7 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 2.9 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.7 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 1.8 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.2 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 1.1 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 8.7 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 2.6 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 3.9 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 2.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 1.9 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 2.1 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 1.6 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 2.5 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 2.9 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.2 | GO:0001093 | TFIIB-class transcription factor binding(GO:0001093) |
| 0.0 | 0.9 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 9.0 | GO:0016491 | oxidoreductase activity(GO:0016491) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 16.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.3 | 22.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 6.7 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 6.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 3.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 1.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 7.4 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 4.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 20.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.8 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.8 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 2.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.6 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 2.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 2.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.6 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 1.1 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 39.2 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.5 | 6.6 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.4 | 4.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.4 | 21.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.3 | 7.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 2.1 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.3 | 4.9 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.2 | 5.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.2 | 2.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 4.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.2 | 11.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 2.9 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 4.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 2.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 2.0 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 2.8 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 2.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 2.8 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.6 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 2.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 2.0 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 8.0 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 0.5 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 0.6 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 2.5 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.1 | 1.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 4.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 3.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.8 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 2.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 2.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.9 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 1.8 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.9 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.8 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.8 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |