Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
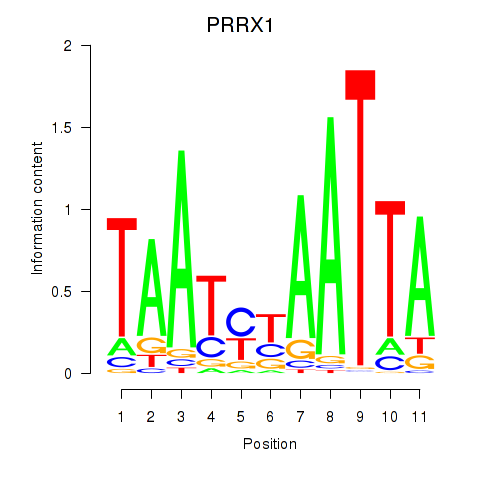
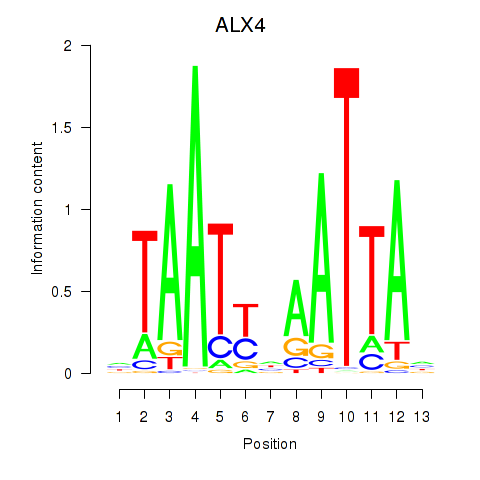
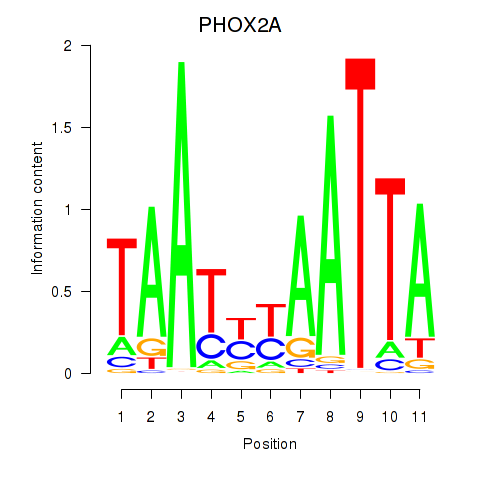
Results for PRRX1_ALX4_PHOX2A
Z-value: 1.54
Transcription factors associated with PRRX1_ALX4_PHOX2A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PRRX1
|
ENSG00000116132.12 | PRRX1 |
|
ALX4
|
ENSG00000052850.8 | ALX4 |
|
PHOX2A
|
ENSG00000165462.5 | PHOX2A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PHOX2A | hg38_v1_chr11_-_72241192_72241275, hg38_v1_chr11_-_72244166_72244184 | -0.12 | 7.4e-02 | Click! |
| PRRX1 | hg38_v1_chr1_+_170664121_170664144 | 0.10 | 1.5e-01 | Click! |
| ALX4 | hg38_v1_chr11_-_44310126_44310152 | 0.09 | 1.8e-01 | Click! |
Activity profile of PRRX1_ALX4_PHOX2A motif
Sorted Z-values of PRRX1_ALX4_PHOX2A motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PRRX1_ALX4_PHOX2A
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 21.9 | 87.5 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 9.6 | 28.9 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 5.5 | 22.0 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 3.6 | 25.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 3.0 | 30.2 | GO:0009449 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 2.8 | 8.5 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 2.5 | 14.8 | GO:0097338 | response to clozapine(GO:0097338) |
| 2.3 | 6.8 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 2.0 | 13.9 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 1.3 | 3.8 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 1.2 | 3.6 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 1.2 | 7.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 1.2 | 4.6 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 1.0 | 3.0 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.9 | 4.6 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.9 | 3.6 | GO:1900191 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.9 | 2.7 | GO:1904440 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.8 | 16.6 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.8 | 35.3 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.8 | 10.2 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.7 | 2.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.7 | 2.6 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.6 | 4.9 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.6 | 4.3 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.6 | 2.4 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.6 | 7.8 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.6 | 4.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.5 | 1.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.5 | 1.6 | GO:0003099 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.5 | 2.3 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.5 | 5.5 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.4 | 2.1 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.4 | 19.0 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.4 | 10.3 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.3 | 1.4 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 0.3 | 59.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.3 | 12.7 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.3 | 1.3 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.3 | 1.3 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.3 | 0.9 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.3 | 0.9 | GO:0061074 | negative regulation of photoreceptor cell differentiation(GO:0046533) regulation of neural retina development(GO:0061074) regulation of retina development in camera-type eye(GO:1902866) |
| 0.3 | 8.6 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.3 | 8.3 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.3 | 2.9 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.3 | 0.8 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.3 | 3.5 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.2 | 1.7 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.2 | 1.8 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.2 | 1.4 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.2 | 16.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.2 | 0.6 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.2 | 4.1 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.2 | 1.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.2 | 0.8 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.2 | 2.0 | GO:2000795 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.2 | 6.0 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.2 | 4.9 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 2.9 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.2 | 13.9 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.2 | 12.8 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.2 | 5.0 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.4 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.1 | 2.0 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 0.4 | GO:1901253 | regulation of intracellular transport of viral material(GO:1901252) negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.1 | 0.7 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.1 | 1.8 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 5.6 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.1 | 1.0 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 2.0 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 0.3 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.1 | 0.7 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.1 | 0.4 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.8 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.1 | 6.5 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.1 | 1.0 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.1 | 5.3 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 2.2 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 1.0 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.1 | 4.0 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 6.1 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 0.9 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.6 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 8.1 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.1 | 1.2 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.1 | 3.6 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.9 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 5.5 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.1 | 4.0 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.1 | 1.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 5.9 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.1 | 0.3 | GO:0021764 | amygdala development(GO:0021764) |
| 0.1 | 1.3 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.1 | 0.2 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.1 | 4.2 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.1 | 1.5 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 2.3 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 | 1.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 13.1 | GO:0060828 | regulation of canonical Wnt signaling pathway(GO:0060828) |
| 0.0 | 1.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.6 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 0.2 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 1.9 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.9 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 7.3 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.0 | 5.3 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.2 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.4 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.0 | 0.1 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.0 | 6.3 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 0.2 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 9.2 | GO:0019216 | regulation of lipid metabolic process(GO:0019216) |
| 0.0 | 1.6 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.2 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.9 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 1.3 | GO:0031016 | pancreas development(GO:0031016) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 87.5 | GO:0005922 | connexon complex(GO:0005922) |
| 3.5 | 14.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 2.3 | 6.8 | GO:0098831 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) presynaptic cytoskeleton(GO:0099569) |
| 1.5 | 36.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 1.3 | 3.8 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 1.2 | 3.6 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.8 | 4.6 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.7 | 7.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.7 | 14.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.6 | 7.8 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.6 | 11.9 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.6 | 4.6 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.5 | 5.5 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.4 | 2.7 | GO:1990357 | terminal web(GO:1990357) |
| 0.3 | 4.0 | GO:0008091 | spectrin(GO:0008091) |
| 0.3 | 5.1 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.3 | 1.2 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.2 | 0.7 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.2 | 22.0 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.2 | 9.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 1.4 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.2 | 5.0 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.2 | 2.0 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 1.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 2.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 1.7 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 0.6 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 18.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 4.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 5.5 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 2.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 59.1 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 7.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 4.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 14.1 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.1 | 22.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 1.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 1.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 2.0 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 9.7 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 0.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 4.1 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.8 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 5.9 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 6.0 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 1.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 1.4 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 1.7 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 2.5 | GO:0044297 | cell body(GO:0044297) |
| 0.0 | 1.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.2 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 1.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 4.3 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 16.0 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.3 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 17.4 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.6 | 87.5 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 3.8 | 11.3 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 2.2 | 30.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 1.2 | 3.6 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 1.1 | 4.3 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 1.0 | 4.0 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.9 | 3.7 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.9 | 14.8 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.9 | 22.0 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.7 | 28.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.5 | 1.6 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.5 | 19.1 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.5 | 13.9 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.5 | 1.5 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.5 | 20.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.5 | 7.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.5 | 5.0 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.5 | 3.6 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.4 | 12.0 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.4 | 5.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.4 | 7.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.4 | 5.5 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.4 | 4.6 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.4 | 4.9 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.4 | 3.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.3 | 5.6 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.3 | 11.8 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.3 | 6.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.3 | 14.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.3 | 8.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.3 | 6.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.3 | 1.2 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.3 | 7.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.3 | 36.4 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.3 | 12.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 2.7 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.2 | 2.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.2 | 4.1 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 1.0 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.2 | 1.4 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 1.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 6.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 2.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.2 | 5.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 4.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 6.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 13.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 2.0 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 1.6 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 14.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 0.3 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.1 | 1.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 17.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 7.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 2.2 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 11.0 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.1 | 0.9 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 23.3 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 4.9 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 2.0 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 1.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.6 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.9 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 54.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 0.6 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 0.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 1.0 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 1.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.2 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.0 | 0.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.8 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 8.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.5 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 1.8 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 3.8 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 88.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.3 | 14.8 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.2 | 13.9 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.2 | 7.3 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.2 | 12.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 5.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 9.0 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 6.8 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 5.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 5.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 6.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 3.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 6.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 7.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 3.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 6.0 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 1.9 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 2.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 2.1 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 6.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 2.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 3.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 10.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.9 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 5.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.9 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 87.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.7 | 22.0 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.5 | 20.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.5 | 14.7 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.4 | 29.5 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.3 | 4.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.3 | 8.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.3 | 7.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 3.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 3.8 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.2 | 5.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.2 | 4.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 8.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 5.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 4.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 5.1 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.1 | 2.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 5.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 2.0 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 11.1 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 10.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 2.0 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 4.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.9 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 0.9 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 1.6 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 1.9 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 6.2 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.7 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 2.2 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |