Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
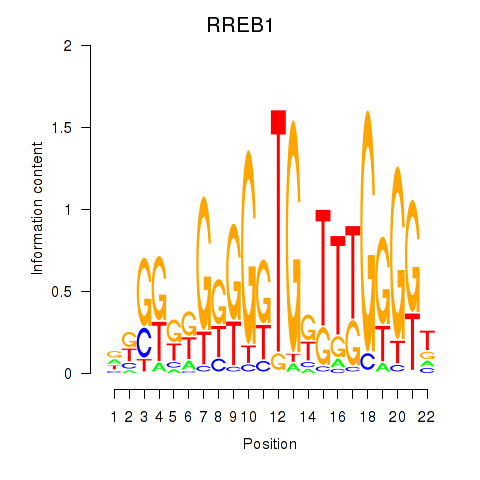
Results for RREB1
Z-value: 3.26
Transcription factors associated with RREB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RREB1
|
ENSG00000124782.20 | RREB1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RREB1 | hg38_v1_chr6_+_7107597_7107821 | 0.40 | 7.4e-10 | Click! |
Activity profile of RREB1 motif
Sorted Z-values of RREB1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RREB1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.6 | GO:0002445 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) type II hypersensitivity(GO:0002445) regulation of type II hypersensitivity(GO:0002892) |
| 2.2 | 8.6 | GO:2000910 | regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 1.7 | 15.5 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 1.7 | 5.0 | GO:0014813 | skeletal muscle satellite cell commitment(GO:0014813) |
| 1.4 | 4.2 | GO:0071988 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 1.3 | 5.2 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 1.2 | 4.9 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 1.1 | 20.1 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 1.0 | 3.0 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 1.0 | 2.0 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.9 | 2.7 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.9 | 17.5 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.9 | 2.6 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.8 | 5.0 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.8 | 3.9 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.8 | 3.0 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.7 | 2.2 | GO:0060032 | notochord regression(GO:0060032) |
| 0.7 | 2.9 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.7 | 2.0 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.6 | 1.9 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.6 | 5.8 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.6 | 3.4 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.6 | 2.2 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.5 | 5.3 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.5 | 1.6 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.5 | 2.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.5 | 5.1 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.5 | 1.5 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.5 | 1.8 | GO:1990834 | negative regulation of T-helper 1 cell differentiation(GO:0045626) response to odorant(GO:1990834) |
| 0.4 | 2.7 | GO:0050893 | sensory processing(GO:0050893) |
| 0.4 | 1.8 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.4 | 3.5 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.4 | 5.9 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.4 | 2.1 | GO:1904501 | glial cell fate determination(GO:0007403) oviduct development(GO:0060066) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) renal inner medulla development(GO:0072053) renal outer medulla development(GO:0072054) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.4 | 1.2 | GO:1904907 | negative regulation of maintenance of sister chromatid cohesion(GO:0034092) negative regulation of maintenance of mitotic sister chromatid cohesion(GO:0034183) maintenance of mitotic sister chromatid cohesion, telomeric(GO:0099403) mitotic sister chromatid cohesion, telomeric(GO:0099404) regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904907) negative regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904908) |
| 0.4 | 1.2 | GO:1905075 | occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.4 | 1.2 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.4 | 5.8 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.4 | 1.9 | GO:0002865 | negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) |
| 0.4 | 4.7 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.4 | 2.2 | GO:0010734 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.4 | 1.1 | GO:0061074 | regulation of neural retina development(GO:0061074) regulation of retina development in camera-type eye(GO:1902866) |
| 0.4 | 7.5 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.3 | 2.1 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.3 | 3.9 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.3 | 4.8 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.3 | 8.4 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.3 | 1.9 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.3 | 3.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.3 | 0.9 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.3 | 5.9 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.3 | 0.8 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.3 | 0.8 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.3 | 0.8 | GO:0051459 | regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.3 | 4.2 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.3 | 4.4 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.3 | 3.3 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.3 | 1.3 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.2 | 1.7 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.2 | 1.9 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.2 | 1.6 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.2 | 5.7 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.2 | 1.3 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.2 | 4.6 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.2 | 0.8 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.2 | 1.3 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.2 | 1.6 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.2 | 2.0 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.2 | 4.0 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.2 | 2.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.2 | 5.2 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.2 | 4.6 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.2 | 3.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 5.1 | GO:0050718 | positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.2 | 2.1 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.2 | 1.0 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.2 | 0.5 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.2 | 1.4 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.2 | 1.7 | GO:0002775 | antimicrobial peptide production(GO:0002775) antibacterial peptide production(GO:0002778) |
| 0.2 | 2.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 0.7 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 1.5 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 2.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 1.0 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.6 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 1.3 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.1 | 10.1 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 0.6 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.1 | 0.5 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 2.0 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 0.2 | GO:0046222 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.1 | 2.4 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 1.3 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 1.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 1.7 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.1 | 0.7 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 4.1 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 | 0.7 | GO:0043697 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.1 | 0.4 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.1 | 1.4 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 1.1 | GO:0036155 | positive regulation of triglyceride catabolic process(GO:0010898) acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.1 | 2.1 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 2.4 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.1 | 4.9 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.1 | 2.5 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.7 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.1 | 0.9 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 0.6 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 | 0.6 | GO:2000322 | regulation of glucocorticoid receptor signaling pathway(GO:2000322) negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.1 | 0.2 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.1 | 0.2 | GO:0031446 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) positive regulation of endoplasmic reticulum calcium ion concentration(GO:0032470) |
| 0.1 | 1.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.4 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 0.4 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.1 | 2.4 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 | 0.5 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 0.6 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 2.5 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.1 | 1.4 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.6 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.1 | 5.3 | GO:0005977 | glycogen metabolic process(GO:0005977) |
| 0.0 | 0.2 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 3.4 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 0.2 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.0 | 1.4 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.2 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.0 | 2.9 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 5.8 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 4.4 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 2.3 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 1.9 | GO:0032210 | regulation of telomere maintenance via telomerase(GO:0032210) |
| 0.0 | 1.1 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.0 | 0.7 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 2.7 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.5 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.0 | 2.0 | GO:0051058 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 1.6 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 1.9 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 1.7 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 0.6 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 1.8 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 4.6 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 1.1 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 1.2 | GO:0006730 | one-carbon metabolic process(GO:0006730) bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:0034311 | sphingosine metabolic process(GO:0006670) diol metabolic process(GO:0034311) |
| 0.0 | 1.3 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 1.4 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 2.5 | GO:0001889 | liver development(GO:0001889) |
| 0.0 | 0.3 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.8 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.7 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.3 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 2.2 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 1.6 | GO:0032479 | regulation of type I interferon production(GO:0032479) |
| 0.0 | 1.1 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.0 | 0.9 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.9 | GO:0030819 | positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.0 | 0.4 | GO:0001755 | neural crest cell migration(GO:0001755) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0070701 | mucus layer(GO:0070701) |
| 1.4 | 10.1 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 1.4 | 4.2 | GO:0055028 | cortical microtubule(GO:0055028) mitotic spindle astral microtubule(GO:0061673) |
| 1.1 | 3.4 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.7 | 2.9 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.7 | 13.9 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.6 | 2.9 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.5 | 8.6 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.5 | 2.9 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.5 | 7.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.4 | 2.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.4 | 1.2 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.4 | 1.9 | GO:0032807 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.3 | 2.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.3 | 3.5 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.3 | 3.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.3 | 2.0 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.3 | 4.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.2 | 3.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 3.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 3.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.2 | 2.9 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 7.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.2 | 2.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 9.8 | GO:0008305 | integrin complex(GO:0008305) |
| 0.2 | 0.7 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.2 | 3.9 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.2 | 1.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.2 | 17.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 6.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 1.7 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.1 | 5.0 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 2.1 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 3.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 2.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.7 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.1 | 24.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 18.9 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.1 | 2.0 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.8 | GO:0051286 | cell tip(GO:0051286) |
| 0.1 | 4.0 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.1 | 1.8 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.1 | 1.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.6 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 4.9 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 2.1 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 1.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.2 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 2.5 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 1.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 2.0 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.6 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 1.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 1.3 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 1.3 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 3.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 7.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 5.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 3.9 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 2.6 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 11.1 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.0 | 2.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 3.0 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 2.3 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 0.3 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 4.8 | GO:0031253 | cell projection membrane(GO:0031253) |
| 0.0 | 1.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 5.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.2 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 6.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 7.3 | GO:0000228 | nuclear chromosome(GO:0000228) |
| 0.0 | 4.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.7 | GO:1904724 | tertiary granule lumen(GO:1904724) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 20.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 1.7 | 8.6 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 1.7 | 5.1 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 1.3 | 4.0 | GO:0032427 | GBD domain binding(GO:0032427) |
| 1.1 | 17.5 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 1.0 | 5.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.9 | 4.4 | GO:0004803 | transposase activity(GO:0004803) |
| 0.9 | 2.6 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.8 | 2.3 | GO:0030305 | heparanase activity(GO:0030305) |
| 0.7 | 2.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.6 | 1.9 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.6 | 4.7 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.6 | 5.2 | GO:0089720 | caspase binding(GO:0089720) |
| 0.5 | 2.7 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.5 | 5.8 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.5 | 5.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.5 | 2.9 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.4 | 3.5 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.4 | 4.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.4 | 1.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.4 | 1.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.4 | 1.2 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) |
| 0.4 | 5.0 | GO:0016918 | retinal binding(GO:0016918) retinol binding(GO:0019841) |
| 0.4 | 3.3 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.4 | 4.8 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.4 | 0.7 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.4 | 2.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.3 | 1.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.3 | 2.2 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.3 | 1.2 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.3 | 3.9 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.3 | 1.6 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.3 | 1.6 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.3 | 1.3 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.2 | 3.0 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.2 | 2.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.2 | 6.5 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.2 | 6.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 1.9 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 1.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 1.7 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 3.9 | GO:0030297 | ErbB-2 class receptor binding(GO:0005176) transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.2 | 3.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.2 | 1.3 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.2 | 1.5 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 7.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.2 | 2.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.2 | 1.5 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 3.1 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.2 | 1.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.7 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 10.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 4.6 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 0.7 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 1.9 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 1.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 2.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 3.7 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 5.1 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 4.8 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.1 | 0.5 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 16.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.2 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 0.3 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.1 | 1.7 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 3.0 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 2.9 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 2.0 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 2.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.6 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.8 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.1 | 0.9 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.1 | 2.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 2.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 1.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 1.5 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 2.0 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 3.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.7 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 1.6 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 1.9 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 2.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.9 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.1 | 2.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 1.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 1.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 1.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 0.8 | GO:0042577 | phosphatidate phosphatase activity(GO:0008195) lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 1.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.5 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 1.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 5.0 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 3.1 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 2.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 2.4 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 2.0 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 10.9 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.9 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 3.3 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.7 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 6.5 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 2.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 3.1 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 7.2 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 6.5 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 20.5 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 3.4 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 3.0 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.7 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.6 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.3 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 2.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.4 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.2 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 10.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 9.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 4.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.2 | 1.8 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 5.6 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.1 | 5.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 7.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 1.3 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 4.9 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 4.8 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 4.0 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 5.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 2.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.7 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 2.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 5.0 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 1.8 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 1.9 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 0.9 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 2.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 3.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 2.9 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 9.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 1.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 3.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 2.9 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.7 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 1.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 4.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.0 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.2 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.5 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.5 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 5.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.7 | 14.2 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.5 | 17.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.4 | 8.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 4.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 5.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 5.1 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 1.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.2 | 3.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 5.3 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 2.6 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 8.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 5.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 6.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.9 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 3.9 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 2.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 2.9 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 1.6 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 3.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 1.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 2.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 2.0 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 0.9 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 0.8 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 2.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 0.7 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 3.0 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 0.4 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 4.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.3 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.7 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 2.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 1.4 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 6.5 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 1.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.7 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |