Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
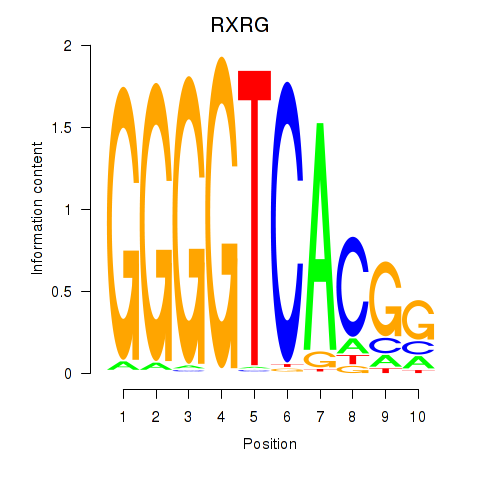
Results for RXRG
Z-value: 6.89
Transcription factors associated with RXRG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RXRG
|
ENSG00000143171.13 | RXRG |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RXRG | hg38_v1_chr1_-_165445220_165445355, hg38_v1_chr1_-_165445088_165445140 | 0.15 | 3.0e-02 | Click! |
Activity profile of RXRG motif
Sorted Z-values of RXRG motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RXRG
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.8 | 23.3 | GO:1900226 | positive regulation of oocyte maturation(GO:1900195) negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 5.6 | 22.5 | GO:0035627 | ceramide transport(GO:0035627) |
| 4.8 | 28.5 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 3.8 | 11.5 | GO:0002884 | negative regulation of hypersensitivity(GO:0002884) |
| 3.6 | 25.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 3.0 | 11.8 | GO:1903294 | regulation of glutamate secretion, neurotransmission(GO:1903294) positive regulation of glutamate secretion, neurotransmission(GO:1903296) |
| 2.6 | 13.0 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 2.5 | 9.9 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 2.5 | 34.3 | GO:0070445 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 2.4 | 7.3 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 2.4 | 9.6 | GO:2000077 | UDP-glucose catabolic process(GO:0006258) positive regulation of adrenergic receptor signaling pathway(GO:0071879) negative regulation of type B pancreatic cell development(GO:2000077) |
| 2.4 | 7.1 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 2.4 | 14.2 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 2.0 | 6.0 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 2.0 | 7.9 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 2.0 | 5.9 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 2.0 | 5.9 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 1.9 | 11.6 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 1.9 | 5.8 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 1.9 | 5.7 | GO:0008355 | olfactory learning(GO:0008355) cellular alkene metabolic process(GO:0043449) |
| 1.9 | 7.5 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 1.9 | 11.3 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 1.8 | 9.2 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 1.8 | 12.7 | GO:0090027 | negative regulation of monocyte chemotaxis(GO:0090027) |
| 1.8 | 7.1 | GO:2000327 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 1.8 | 10.6 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 1.7 | 10.4 | GO:0097460 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 1.7 | 5.2 | GO:0051232 | meiotic spindle elongation(GO:0051232) meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 1.7 | 6.8 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) late viral transcription(GO:0019086) |
| 1.7 | 6.6 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 1.7 | 9.9 | GO:0016926 | protein desumoylation(GO:0016926) |
| 1.6 | 4.7 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 1.5 | 4.6 | GO:0099541 | trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 1.5 | 7.7 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 1.5 | 4.6 | GO:0060031 | mediolateral intercalation(GO:0060031) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) |
| 1.5 | 10.2 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 1.4 | 5.8 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 1.4 | 4.1 | GO:0034092 | negative regulation of maintenance of sister chromatid cohesion(GO:0034092) negative regulation of maintenance of mitotic sister chromatid cohesion(GO:0034183) maintenance of mitotic sister chromatid cohesion, telomeric(GO:0099403) mitotic sister chromatid cohesion, telomeric(GO:0099404) regulation of telomeric DNA binding(GO:1904742) regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904907) negative regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904908) |
| 1.3 | 6.6 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 1.3 | 3.9 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 1.3 | 12.8 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 1.3 | 3.8 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 1.3 | 3.8 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 1.3 | 6.3 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 1.2 | 11.2 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 1.2 | 6.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 1.2 | 12.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 1.2 | 8.4 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 1.2 | 7.1 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 1.1 | 5.7 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 1.1 | 4.6 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 1.1 | 23.8 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 1.1 | 4.4 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 1.1 | 4.4 | GO:1902617 | response to fluoride(GO:1902617) |
| 1.0 | 9.3 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 1.0 | 3.1 | GO:0033082 | extrathymic T cell differentiation(GO:0033078) regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 1.0 | 3.1 | GO:0009720 | detection of hormone stimulus(GO:0009720) |
| 1.0 | 22.3 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 1.0 | 3.0 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 1.0 | 11.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 1.0 | 3.0 | GO:0071529 | cementum mineralization(GO:0071529) |
| 1.0 | 3.9 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 1.0 | 2.9 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.9 | 3.7 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.9 | 6.2 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.9 | 8.9 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.9 | 4.4 | GO:0019732 | antifungal humoral response(GO:0019732) |
| 0.9 | 4.3 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.8 | 2.5 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.8 | 7.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.8 | 4.1 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.8 | 6.3 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.8 | 1.6 | GO:0032242 | regulation of nucleoside transport(GO:0032242) |
| 0.8 | 2.3 | GO:0035565 | regulation of pronephros size(GO:0035565) |
| 0.8 | 6.8 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.8 | 15.9 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.8 | 2.3 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.7 | 3.5 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.7 | 2.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.7 | 0.7 | GO:1990641 | response to iron ion starvation(GO:1990641) |
| 0.7 | 4.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.7 | 2.7 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.7 | 2.0 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) |
| 0.7 | 11.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.7 | 8.5 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.7 | 2.0 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) regulation of positive thymic T cell selection(GO:1902232) |
| 0.6 | 2.6 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.6 | 7.7 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.6 | 1.8 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.6 | 8.0 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.6 | 4.9 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.6 | 10.9 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.6 | 9.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.6 | 7.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.6 | 4.7 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.6 | 5.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.6 | 5.8 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.6 | 5.2 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.6 | 2.9 | GO:0070778 | L-aspartate transport(GO:0070778) L-aspartate transmembrane transport(GO:0089712) |
| 0.6 | 5.7 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.6 | 1.7 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.6 | 5.5 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.6 | 2.2 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.5 | 2.2 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.5 | 6.5 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.5 | 4.7 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.5 | 5.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.5 | 3.6 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.5 | 3.0 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.5 | 2.0 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.5 | 2.0 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.5 | 3.5 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.5 | 4.5 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.5 | 7.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.5 | 12.4 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.5 | 2.5 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.5 | 8.9 | GO:0008212 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.5 | 28.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.5 | 4.4 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.5 | 5.9 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.5 | 3.9 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.5 | 3.9 | GO:1904995 | negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.5 | 5.7 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.5 | 2.3 | GO:1904075 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.5 | 3.3 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.5 | 2.3 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.5 | 10.1 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.5 | 3.6 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.5 | 3.2 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.5 | 3.2 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.4 | 4.8 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.4 | 2.5 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.4 | 1.3 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.4 | 0.8 | GO:1902612 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.4 | 6.6 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.4 | 13.6 | GO:0042749 | regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.4 | 4.9 | GO:0046958 | nonassociative learning(GO:0046958) |
| 0.4 | 1.6 | GO:0035962 | response to interleukin-13(GO:0035962) |
| 0.4 | 4.5 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.4 | 13.4 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.4 | 2.8 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.4 | 1.2 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.4 | 5.8 | GO:0030903 | notochord development(GO:0030903) |
| 0.4 | 2.7 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.4 | 3.1 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.4 | 2.3 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.4 | 5.0 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.4 | 4.5 | GO:0021794 | thalamus development(GO:0021794) |
| 0.4 | 4.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.4 | 3.4 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.4 | 1.1 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.4 | 4.1 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.4 | 1.5 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.4 | 5.9 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.4 | 1.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.4 | 1.8 | GO:0048861 | oncostatin-M-mediated signaling pathway(GO:0038165) leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.4 | 5.0 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.3 | 8.8 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.3 | 3.0 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.3 | 1.3 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.3 | 2.9 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.3 | 0.6 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.3 | 1.3 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.3 | 2.5 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.3 | 2.5 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.3 | 1.5 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.3 | 3.6 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.3 | 0.9 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) |
| 0.3 | 2.1 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.3 | 11.3 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.3 | 28.5 | GO:0006073 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.3 | 2.3 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.3 | 2.3 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.3 | 5.5 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.3 | 3.4 | GO:0048243 | norepinephrine secretion(GO:0048243) |
| 0.3 | 1.7 | GO:0061205 | alveolar primary septum development(GO:0061143) paramesonephric duct development(GO:0061205) |
| 0.3 | 2.5 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.3 | 2.5 | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) |
| 0.3 | 4.1 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.3 | 3.2 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.3 | 3.1 | GO:0042635 | positive regulation of hair cycle(GO:0042635) positive regulation of hair follicle development(GO:0051798) |
| 0.3 | 13.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.3 | 0.5 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.3 | 3.4 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.3 | 4.3 | GO:0090344 | negative regulation of cell aging(GO:0090344) |
| 0.3 | 7.9 | GO:0003416 | endochondral bone growth(GO:0003416) tongue development(GO:0043586) |
| 0.2 | 7.9 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.2 | 12.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 3.0 | GO:0045794 | negative regulation of cell volume(GO:0045794) |
| 0.2 | 4.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 3.6 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.2 | 5.1 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.2 | 1.9 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) |
| 0.2 | 2.1 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.2 | 1.7 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.2 | 0.7 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.2 | 3.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 2.0 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.2 | 3.3 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.2 | 4.8 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.2 | 3.5 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.2 | 1.3 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.2 | 1.5 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.2 | 3.8 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.2 | 3.6 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.2 | 1.5 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.2 | 0.8 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.2 | 3.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.2 | 0.8 | GO:1990418 | response to sodium phosphate(GO:1904383) response to insulin-like growth factor stimulus(GO:1990418) |
| 0.2 | 1.8 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.2 | 3.2 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.2 | 5.0 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.2 | 1.8 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 2.3 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.2 | 2.0 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.2 | 8.2 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.2 | 6.0 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.2 | 7.3 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.2 | 3.1 | GO:0019068 | virion assembly(GO:0019068) |
| 0.2 | 3.1 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.2 | 7.1 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.2 | 1.0 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.2 | 1.5 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.2 | 0.5 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.2 | 3.9 | GO:1900273 | positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.2 | 4.7 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.2 | 2.8 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.2 | 1.6 | GO:0007614 | short-term memory(GO:0007614) |
| 0.2 | 3.0 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.1 | 4.3 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 2.5 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.1 | 5.6 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 4.5 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.1 | 1.6 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 3.8 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.1 | 3.9 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 4.2 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 1.6 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.1 | 2.9 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 1.2 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.1 | 1.3 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 | 5.4 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 4.0 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 2.5 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 5.5 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.1 | 3.3 | GO:0014898 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.1 | 1.6 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.1 | 0.4 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 2.3 | GO:0003170 | heart valve development(GO:0003170) |
| 0.1 | 2.8 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.1 | 2.3 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 | 2.3 | GO:0009798 | axis specification(GO:0009798) |
| 0.1 | 2.3 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 5.6 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 4.6 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.1 | 4.8 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 2.3 | GO:0009081 | branched-chain amino acid metabolic process(GO:0009081) branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 1.3 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 2.0 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.1 | 0.5 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 2.0 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 2.0 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 1.7 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.1 | 1.6 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 1.0 | GO:0007616 | long-term memory(GO:0007616) |
| 0.1 | 5.0 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.1 | 2.4 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.1 | 2.2 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 3.3 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.1 | 2.4 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.1 | 0.8 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 1.8 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.1 | 0.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 3.6 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 1.5 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 1.4 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 3.2 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 1.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 4.1 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.5 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 1.2 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.9 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.6 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 2.0 | GO:0018022 | peptidyl-lysine methylation(GO:0018022) |
| 0.0 | 5.8 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 5.1 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.0 | 1.1 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 1.0 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 2.2 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 2.1 | GO:0060996 | dendritic spine development(GO:0060996) |
| 0.0 | 2.0 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.9 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.4 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.8 | GO:0045453 | bone resorption(GO:0045453) |
| 0.0 | 0.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.8 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.0 | 0.1 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.9 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 29.7 | GO:0072534 | perineuronal net(GO:0072534) |
| 3.1 | 12.4 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 2.6 | 23.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 2.2 | 8.9 | GO:0036128 | CatSper complex(GO:0036128) |
| 2.0 | 7.9 | GO:0048179 | activin receptor complex(GO:0048179) |
| 2.0 | 11.8 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 2.0 | 5.9 | GO:0098831 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) presynaptic cytoskeleton(GO:0099569) |
| 1.9 | 18.8 | GO:0070852 | cell body fiber(GO:0070852) |
| 1.8 | 19.6 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 1.7 | 10.4 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 1.5 | 13.8 | GO:0033391 | chromatoid body(GO:0033391) |
| 1.5 | 7.3 | GO:0097513 | myosin II filament(GO:0097513) |
| 1.5 | 16.0 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 1.3 | 6.3 | GO:0045160 | myosin I complex(GO:0045160) |
| 1.2 | 6.2 | GO:0036398 | TCR signalosome(GO:0036398) |
| 1.1 | 4.5 | GO:0000801 | central element(GO:0000801) |
| 1.1 | 15.5 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 1.1 | 15.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 1.0 | 2.9 | GO:0060987 | lipid tube(GO:0060987) |
| 0.9 | 6.6 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.9 | 4.7 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.9 | 4.6 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.9 | 1.8 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.9 | 17.9 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.9 | 6.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.9 | 4.4 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.9 | 2.6 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.8 | 7.5 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.8 | 5.8 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.8 | 17.0 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.8 | 13.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.8 | 14.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.8 | 10.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.7 | 2.0 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.7 | 14.1 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.7 | 4.0 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.6 | 7.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.6 | 11.5 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.6 | 5.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.6 | 3.9 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.5 | 6.4 | GO:0071203 | WASH complex(GO:0071203) |
| 0.5 | 4.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.5 | 6.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.4 | 3.0 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.4 | 9.0 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.4 | 5.7 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.4 | 2.3 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.4 | 4.6 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.4 | 4.8 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.4 | 7.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.4 | 1.8 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.4 | 12.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.3 | 7.4 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.3 | 3.0 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.3 | 6.3 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.3 | 2.3 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.3 | 4.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.3 | 3.7 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.3 | 1.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.3 | 4.0 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.3 | 17.5 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.2 | 7.5 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.2 | 2.5 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.2 | 3.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.2 | 12.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.2 | 2.0 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.2 | 18.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.2 | 2.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 1.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.2 | 0.4 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.2 | 1.5 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.2 | 5.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 7.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 3.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.2 | 2.0 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 2.1 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 16.2 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.2 | 1.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.2 | 2.5 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.2 | 5.0 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 6.1 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 3.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 4.1 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 9.5 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 1.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 3.8 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 1.6 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 1.7 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 4.1 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 3.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 1.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 26.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 7.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 11.9 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 7.8 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 1.6 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.1 | 4.4 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 2.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 7.6 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 1.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 4.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 3.5 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.9 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 12.7 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 1.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 4.3 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 3.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 5.7 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 4.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 6.8 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.1 | 2.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 5.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 4.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 11.7 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 2.7 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 1.1 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 3.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 11.2 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.9 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 1.0 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 1.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.7 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.5 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 1.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 2.6 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.8 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 1.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 2.5 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 1.1 | GO:0005796 | Golgi lumen(GO:0005796) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.8 | 23.3 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 5.9 | 23.7 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 5.6 | 22.5 | GO:0097001 | ceramide binding(GO:0097001) |
| 5.2 | 15.5 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 4.1 | 20.5 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 3.6 | 25.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 3.5 | 28.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 2.5 | 12.7 | GO:0004803 | transposase activity(GO:0004803) |
| 2.1 | 6.3 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 2.0 | 6.0 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 2.0 | 15.6 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 1.8 | 11.1 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 1.8 | 9.2 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 1.8 | 10.7 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 1.7 | 10.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 1.7 | 9.9 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 1.5 | 9.0 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 1.5 | 10.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 1.4 | 9.9 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 1.4 | 9.8 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 1.4 | 8.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 1.4 | 5.6 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 1.4 | 4.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 1.3 | 7.9 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 1.3 | 17.7 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 1.2 | 9.8 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 1.2 | 7.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 1.2 | 5.8 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 1.1 | 1.1 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 1.1 | 4.4 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 1.1 | 11.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 1.0 | 9.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 1.0 | 4.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 1.0 | 10.6 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 1.0 | 2.9 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 1.0 | 2.9 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 1.0 | 3.8 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.9 | 8.5 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.9 | 4.7 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.9 | 2.7 | GO:0004507 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.9 | 28.9 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.9 | 4.5 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.9 | 4.4 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.8 | 6.6 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.8 | 10.4 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.8 | 33.4 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.8 | 3.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.8 | 10.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.7 | 17.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.7 | 20.1 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.7 | 2.1 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.7 | 18.7 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.7 | 4.6 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.7 | 3.9 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.7 | 4.6 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.6 | 5.8 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.6 | 5.7 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.6 | 8.8 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.6 | 1.9 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.6 | 7.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.6 | 8.0 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.6 | 1.8 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.6 | 4.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.6 | 2.9 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.6 | 7.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.6 | 13.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.6 | 11.8 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.6 | 23.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.5 | 1.6 | GO:0047325 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol tetrakisphosphate kinase activity(GO:0051765) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.5 | 3.8 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.5 | 7.7 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.5 | 1.5 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.5 | 3.5 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.5 | 11.8 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.5 | 9.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.5 | 3.6 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.4 | 3.6 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.4 | 9.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.4 | 3.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.4 | 6.1 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.4 | 1.6 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.4 | 4.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.4 | 1.1 | GO:0098918 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.4 | 6.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.4 | 2.8 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.4 | 2.5 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.3 | 3.5 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.3 | 3.5 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.3 | 3.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.3 | 6.3 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.3 | 2.3 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.3 | 3.0 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.3 | 2.9 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.3 | 4.9 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.3 | 6.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.3 | 1.6 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.3 | 5.0 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.3 | 4.6 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.3 | 4.3 | GO:0046790 | virion binding(GO:0046790) |
| 0.3 | 8.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.3 | 3.0 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.3 | 2.6 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.3 | 0.8 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.3 | 32.1 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.3 | 1.9 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.3 | 12.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.3 | 3.9 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.3 | 4.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 1.3 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.3 | 1.8 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.3 | 19.4 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.3 | 3.8 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 3.0 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.2 | 4.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 1.7 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.2 | 3.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 1.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.2 | 1.4 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.2 | 0.9 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.2 | 1.3 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.2 | 3.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 2.6 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 1.5 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.2 | 0.8 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.2 | 6.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.2 | 10.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 1.9 | GO:0016594 | glycine binding(GO:0016594) |
| 0.2 | 6.9 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.2 | 3.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 5.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.2 | 2.9 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 2.2 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.2 | 17.9 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.2 | 1.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 4.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.2 | 2.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 8.2 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 8.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 1.3 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 0.4 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) rhodopsin kinase activity(GO:0050254) |
| 0.1 | 2.0 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 4.8 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 1.8 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 3.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 5.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.7 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.1 | 2.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 6.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 3.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 1.9 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 9.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 1.1 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.1 | 17.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.9 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 1.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 1.6 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 23.1 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 2.1 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 3.8 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 2.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 3.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 3.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 1.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.5 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 1.9 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 0.6 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 2.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 1.3 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 9.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 1.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 5.5 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 2.1 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 1.7 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.1 | 5.1 | GO:0001159 | core promoter proximal region DNA binding(GO:0001159) |
| 0.1 | 1.3 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 1.0 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.4 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 2.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 5.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 3.3 | GO:0008144 | drug binding(GO:0008144) |
| 0.1 | 2.9 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.1 | 2.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 2.4 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 1.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.5 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 1.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 2.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 11.8 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.4 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 3.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 1.0 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 1.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 7.7 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 0.7 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 2.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.9 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.0 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 27.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.6 | 12.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.5 | 29.1 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.5 | 8.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.5 | 31.8 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.4 | 7.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.4 | 1.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.4 | 10.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.3 | 5.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.3 | 23.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.3 | 7.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.3 | 11.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.3 | 10.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.3 | 8.4 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.3 | 4.9 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.2 | 4.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.2 | 11.1 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 6.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 2.6 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.2 | 14.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 4.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 12.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.2 | 7.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 9.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.2 | 6.8 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.2 | 9.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 7.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 7.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 7.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 7.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 2.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 2.3 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 2.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 4.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 2.2 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 1.8 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 4.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 4.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 3.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 1.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 9.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 5.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 1.6 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 2.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 1.2 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 1.6 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 2.9 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 11.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 5.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.9 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.5 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 11.8 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.8 | 27.0 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.7 | 11.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.6 | 12.9 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.6 | 7.4 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.6 | 12.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.5 | 11.8 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.5 | 17.0 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.5 | 5.9 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.5 | 6.4 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.4 | 13.0 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.4 | 11.0 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.4 | 8.8 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.4 | 8.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.4 | 16.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.4 | 14.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.4 | 5.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.4 | 2.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.4 | 6.7 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.3 | 13.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.3 | 2.0 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.3 | 15.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.3 | 7.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.3 | 12.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.3 | 16.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.3 | 4.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.3 | 4.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.3 | 11.4 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.3 | 7.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.3 | 16.0 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.3 | 6.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.3 | 1.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.3 | 5.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 36.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.2 | 4.2 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.2 | 3.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 5.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 3.0 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.2 | 1.9 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.2 | 6.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 23.4 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.2 | 4.7 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.2 | 5.7 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.2 | 8.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 4.8 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.2 | 4.9 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.2 | 1.6 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.2 | 15.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 3.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.9 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 3.6 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 0.4 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.1 | 7.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 3.7 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 7.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.8 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 2.9 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 1.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 17.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 2.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 3.2 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 2.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.0 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 3.2 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 1.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 4.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 0.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 1.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 0.8 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 1.6 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 15.2 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 2.5 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.4 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 1.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.1 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.2 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.9 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.3 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |