Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for SIX5_SMARCC2_HCFC1
Z-value: 5.71
Transcription factors associated with SIX5_SMARCC2_HCFC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
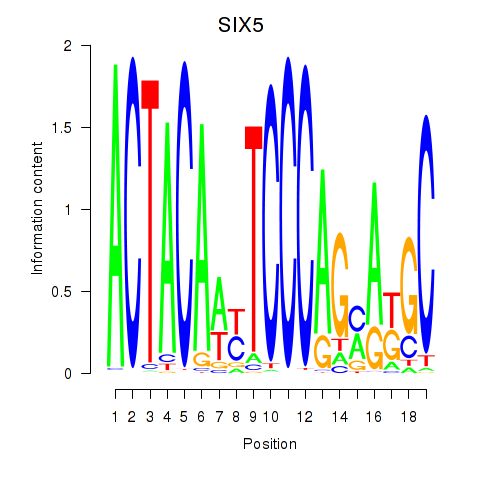
SIX5
|
ENSG00000177045.10 | SIX5 |
|
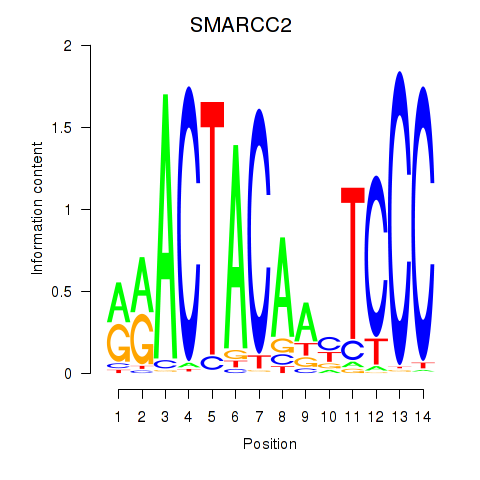
SMARCC2
|
ENSG00000139613.12 | SMARCC2 |
|
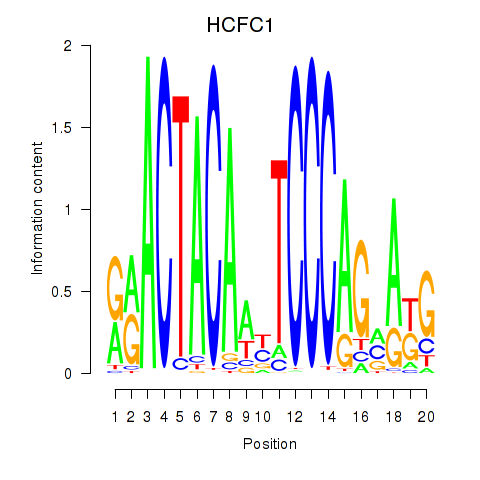
HCFC1
|
ENSG00000172534.14 | HCFC1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HCFC1 | hg38_v1_chrX_-_153971169_153971233, hg38_v1_chrX_-_153971810_153971844 | 0.41 | 4.4e-10 | Click! |
| SMARCC2 | hg38_v1_chr12_-_56189548_56189609 | 0.38 | 5.2e-09 | Click! |
| SIX5 | hg38_v1_chr19_-_45768843_45768869 | -0.37 | 2.5e-08 | Click! |
Activity profile of SIX5_SMARCC2_HCFC1 motif
Sorted Z-values of SIX5_SMARCC2_HCFC1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SIX5_SMARCC2_HCFC1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.5 | 94.3 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 12.4 | 49.4 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 12.3 | 110.3 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 11.4 | 34.2 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 10.9 | 54.7 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 9.3 | 27.8 | GO:1902594 | viral penetration into host nucleus(GO:0075732) multi-organism nuclear import(GO:1902594) |
| 8.9 | 44.3 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 8.8 | 35.1 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 8.7 | 43.6 | GO:0033383 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 8.0 | 39.9 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 7.8 | 39.0 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 7.5 | 52.7 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 6.1 | 18.4 | GO:0070343 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 5.9 | 47.3 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 5.8 | 69.2 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 5.6 | 16.7 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 5.6 | 22.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 5.4 | 38.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 5.3 | 16.0 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 4.9 | 54.0 | GO:0006108 | malate metabolic process(GO:0006108) |
| 4.6 | 59.9 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 4.6 | 13.8 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) protein localization to vacuolar membrane(GO:1903778) |
| 4.6 | 4.6 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 4.4 | 115.2 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 4.3 | 43.5 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 4.3 | 60.6 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 4.2 | 29.2 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 3.9 | 15.8 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 3.9 | 15.7 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 3.5 | 7.1 | GO:0032240 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 3.3 | 16.5 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 3.3 | 13.2 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 3.3 | 13.2 | GO:0036481 | intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036481) |
| 3.3 | 69.0 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 3.3 | 16.3 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 3.3 | 13.0 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 3.2 | 9.7 | GO:0031938 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) regulation of chromatin silencing at telomere(GO:0031938) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 3.1 | 18.9 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 3.1 | 34.2 | GO:0042407 | cristae formation(GO:0042407) |
| 3.1 | 27.8 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 3.0 | 11.9 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 3.0 | 11.9 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 2.8 | 11.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 2.6 | 15.8 | GO:0030581 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 2.6 | 2.6 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 2.5 | 7.6 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 2.5 | 25.1 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 2.5 | 12.5 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) |
| 2.5 | 7.4 | GO:0034471 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 2.5 | 24.7 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 2.5 | 12.3 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 2.4 | 24.3 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 2.4 | 7.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 2.3 | 11.7 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 2.3 | 11.6 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 2.3 | 16.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 2.3 | 9.0 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 2.2 | 13.3 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 2.2 | 17.6 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 2.2 | 13.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 2.1 | 4.2 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 2.1 | 10.4 | GO:0051511 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 2.0 | 16.4 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 1.9 | 17.5 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 1.8 | 12.9 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 1.8 | 8.9 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 1.8 | 35.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 1.8 | 35.3 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 1.7 | 47.7 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 1.6 | 8.2 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 1.6 | 4.9 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 1.6 | 6.4 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 1.5 | 9.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 1.5 | 19.3 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 1.5 | 7.4 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 1.5 | 16.2 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 1.4 | 55.1 | GO:0071174 | mitotic spindle checkpoint(GO:0071174) |
| 1.4 | 5.7 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) peptidyl-glycine modification(GO:0018201) |
| 1.4 | 5.7 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 1.4 | 107.2 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 1.4 | 5.6 | GO:1904636 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 1.4 | 23.7 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 1.4 | 4.1 | GO:0021678 | fourth ventricle development(GO:0021592) third ventricle development(GO:0021678) |
| 1.3 | 4.0 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 1.3 | 3.9 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 1.3 | 5.1 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 1.2 | 14.4 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 1.2 | 21.5 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 1.2 | 7.2 | GO:2001270 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) |
| 1.1 | 19.5 | GO:1902074 | G1 to G0 transition(GO:0070314) response to salt(GO:1902074) |
| 1.1 | 3.4 | GO:0051793 | medium-chain fatty acid catabolic process(GO:0051793) |
| 1.1 | 5.7 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 1.1 | 15.8 | GO:1904871 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 1.1 | 16.4 | GO:0007091 | metaphase/anaphase transition of mitotic cell cycle(GO:0007091) |
| 1.0 | 3.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 1.0 | 12.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 1.0 | 13.9 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 1.0 | 4.8 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.9 | 13.2 | GO:1903441 | protein localization to ciliary membrane(GO:1903441) |
| 0.9 | 8.4 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.9 | 14.8 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.9 | 3.6 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 0.9 | 14.5 | GO:0051974 | negative regulation of telomerase activity(GO:0051974) |
| 0.9 | 19.7 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.9 | 3.5 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.9 | 2.6 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.9 | 4.3 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.8 | 49.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.8 | 12.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.8 | 4.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.8 | 9.7 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.8 | 29.9 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.8 | 2.4 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.8 | 11.0 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.8 | 11.4 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.8 | 12.1 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.8 | 2.3 | GO:0072229 | proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.7 | 16.8 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.7 | 52.3 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.7 | 6.5 | GO:1900223 | positive regulation of beta-amyloid clearance(GO:1900223) |
| 0.7 | 7.2 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.7 | 1.4 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.7 | 2.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.7 | 3.5 | GO:0009213 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) |
| 0.7 | 2.1 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) negative regulation of cell cycle checkpoint(GO:1901977) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.7 | 9.6 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.7 | 2.7 | GO:1902775 | mitochondrial ribosome assembly(GO:0061668) mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.7 | 4.6 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.7 | 16.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.6 | 4.5 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.6 | 5.7 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.6 | 5.7 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.6 | 19.3 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.6 | 3.1 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.6 | 20.8 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.6 | 4.9 | GO:0002176 | male germ cell proliferation(GO:0002176) germ cell proliferation(GO:0036093) |
| 0.6 | 26.3 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.6 | 66.0 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.6 | 8.3 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.6 | 6.5 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.6 | 5.2 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.6 | 2.9 | GO:0052572 | response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 0.6 | 2.9 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.6 | 21.6 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.6 | 11.1 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.5 | 37.4 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.5 | 2.7 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.5 | 16.9 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.5 | 6.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.5 | 3.0 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.5 | 4.0 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.5 | 1.9 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.5 | 1.0 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.5 | 1.4 | GO:0001188 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.5 | 30.2 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.5 | 3.2 | GO:0010624 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.5 | 3.6 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.5 | 4.1 | GO:1905216 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.4 | 0.9 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.4 | 4.0 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.4 | 29.5 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.4 | 3.0 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.4 | 5.0 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.4 | 4.6 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.4 | 5.3 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.4 | 2.4 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.4 | 10.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.4 | 3.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.4 | 4.7 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) DNA integration(GO:0015074) |
| 0.4 | 3.4 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.4 | 10.1 | GO:0001825 | blastocyst formation(GO:0001825) |
| 0.4 | 2.2 | GO:0070200 | establishment of protein localization to telomere(GO:0070200) |
| 0.4 | 2.5 | GO:0044036 | cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.4 | 1.4 | GO:0030047 | actin modification(GO:0030047) |
| 0.4 | 2.8 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.3 | 3.1 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.3 | 1.0 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.3 | 14.7 | GO:0070423 | nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway(GO:0035872) nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) |
| 0.3 | 3.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.3 | 1.3 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.3 | 5.0 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.3 | 2.2 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.3 | 8.0 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.3 | 4.0 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.3 | 3.9 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.3 | 8.7 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.3 | 1.5 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.3 | 1.8 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.3 | 5.2 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.3 | 4.0 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.3 | 3.7 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.3 | 8.8 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.3 | 2.0 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.3 | 1.7 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.3 | 0.8 | GO:0051105 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.3 | 1.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.3 | 2.7 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.3 | 7.3 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.3 | 3.2 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.3 | 3.7 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.2 | 0.7 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.2 | 13.7 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.2 | 2.6 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.2 | 2.8 | GO:0098792 | xenophagy(GO:0098792) |
| 0.2 | 2.3 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.2 | 2.3 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.2 | 6.0 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.2 | 25.6 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.2 | 0.7 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.2 | 13.4 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.2 | 1.8 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.2 | 12.0 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.2 | 4.3 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.2 | 2.6 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.2 | 1.0 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.2 | 3.5 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.2 | 0.8 | GO:1903788 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) negative regulation of vascular associated smooth muscle cell migration(GO:1904753) |
| 0.2 | 7.1 | GO:0006101 | citrate metabolic process(GO:0006101) |
| 0.2 | 1.2 | GO:1904526 | regulation of microtubule binding(GO:1904526) |
| 0.2 | 0.6 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.2 | 5.5 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.2 | 0.8 | GO:0003011 | involuntary skeletal muscle contraction(GO:0003011) |
| 0.2 | 4.7 | GO:1901661 | quinone metabolic process(GO:1901661) |
| 0.2 | 1.3 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.2 | 1.1 | GO:0090114 | COPII-coated vesicle budding(GO:0090114) |
| 0.2 | 2.0 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.2 | 0.4 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.2 | 18.4 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.2 | 5.7 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.2 | 1.5 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.2 | 0.3 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.2 | 2.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 8.6 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.2 | 2.4 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 10.9 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 3.1 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.1 | 4.0 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.1 | 1.1 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 36.3 | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.1 | 0.7 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.1 | 3.0 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.1 | 4.2 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 0.8 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.1 | 0.7 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.8 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.1 | 1.4 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 0.6 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.1 | 1.8 | GO:0097284 | hepatocyte apoptotic process(GO:0097284) |
| 0.1 | 1.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 1.6 | GO:1903286 | regulation of potassium ion import(GO:1903286) |
| 0.1 | 1.9 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 3.6 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 0.6 | GO:0090045 | positive regulation of deacetylase activity(GO:0090045) |
| 0.1 | 3.4 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.1 | 1.6 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 1.8 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 3.0 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.1 | 12.0 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.1 | 0.7 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 2.3 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.1 | 6.3 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 30.6 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.1 | 11.6 | GO:0046546 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.1 | 1.0 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 2.8 | GO:0050766 | positive regulation of phagocytosis(GO:0050766) |
| 0.1 | 2.8 | GO:1990090 | cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.1 | 3.0 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.1 | 2.3 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.1 | 1.3 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.1 | 6.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.7 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.1 | 9.8 | GO:0010952 | positive regulation of peptidase activity(GO:0010952) |
| 0.1 | 1.9 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 0.6 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 3.4 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 5.0 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.1 | 1.6 | GO:0009081 | branched-chain amino acid metabolic process(GO:0009081) branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 0.5 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.1 | 0.8 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 10.6 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.1 | 0.8 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.1 | 6.9 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.1 | 10.7 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.1 | 1.9 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.1 | 1.3 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 | 0.6 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.8 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.1 | GO:0015847 | putrescine transport(GO:0015847) |
| 0.0 | 4.0 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 1.0 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 4.9 | GO:0006606 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) |
| 0.0 | 3.6 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.6 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.7 | GO:0015800 | acidic amino acid transport(GO:0015800) L-glutamate transport(GO:0015813) |
| 0.0 | 0.3 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.2 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 1.5 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.0 | 0.7 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.0 | 0.1 | GO:0046851 | negative regulation of tissue remodeling(GO:0034104) negative regulation of bone resorption(GO:0045779) negative regulation of bone remodeling(GO:0046851) |
| 0.0 | 1.0 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 3.8 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.6 | GO:0006221 | pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.0 | 0.3 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 1.1 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.3 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.4 | GO:0018126 | protein hydroxylation(GO:0018126) |
| 0.0 | 0.1 | GO:0043305 | negative regulation of immunoglobulin production(GO:0002638) negative regulation of mast cell activation involved in immune response(GO:0033007) negative regulation of mast cell degranulation(GO:0043305) |
| 0.0 | 0.2 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 0.2 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.2 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.4 | GO:0009168 | purine nucleoside monophosphate biosynthetic process(GO:0009127) purine ribonucleoside monophosphate biosynthetic process(GO:0009168) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 18.9 | 94.3 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 14.5 | 43.5 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 12.3 | 36.8 | GO:0033597 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 9.5 | 47.7 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 8.5 | 25.6 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 8.2 | 49.0 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 7.5 | 52.8 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 7.5 | 44.7 | GO:0002177 | manchette(GO:0002177) |
| 7.1 | 42.9 | GO:0061617 | MICOS complex(GO:0061617) |
| 6.1 | 60.6 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 5.8 | 64.3 | GO:0070449 | elongin complex(GO:0070449) |
| 5.6 | 33.7 | GO:0001940 | male pronucleus(GO:0001940) |
| 5.2 | 83.9 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 5.1 | 112.7 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 5.1 | 30.4 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 4.9 | 126.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 4.4 | 13.3 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 4.4 | 17.6 | GO:0071986 | Ragulator complex(GO:0071986) |
| 4.2 | 54.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 3.9 | 15.8 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 3.7 | 11.0 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 3.7 | 33.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 3.6 | 10.7 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 3.5 | 39.0 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 3.5 | 21.0 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 3.5 | 13.9 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 3.4 | 13.7 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 3.2 | 9.7 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 3.0 | 29.8 | GO:0070552 | BRISC complex(GO:0070552) |
| 2.9 | 14.3 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 2.8 | 14.2 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 2.8 | 8.4 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 2.8 | 11.1 | GO:0035363 | histone locus body(GO:0035363) |
| 2.7 | 16.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 2.6 | 7.9 | GO:0030689 | Noc complex(GO:0030689) |
| 2.6 | 23.7 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 2.5 | 35.5 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 2.4 | 29.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 2.4 | 19.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 2.4 | 7.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 2.3 | 29.5 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 2.3 | 9.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 2.2 | 6.5 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 2.1 | 8.2 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 2.0 | 12.1 | GO:0005816 | spindle pole body(GO:0005816) |
| 2.0 | 34.1 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 1.9 | 52.2 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 1.9 | 21.0 | GO:0005638 | lamin filament(GO:0005638) |
| 1.8 | 16.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 1.8 | 17.8 | GO:0043203 | axon hillock(GO:0043203) |
| 1.7 | 24.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 1.7 | 8.5 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 1.7 | 67.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 1.6 | 4.9 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 1.6 | 30.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 1.5 | 29.1 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 1.5 | 4.6 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 1.5 | 16.6 | GO:0005686 | U2 snRNP(GO:0005686) |
| 1.4 | 9.6 | GO:1990130 | Iml1 complex(GO:1990130) |
| 1.4 | 13.6 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 1.3 | 8.9 | GO:0005687 | U4 snRNP(GO:0005687) |
| 1.2 | 8.7 | GO:0016272 | prefoldin complex(GO:0016272) |
| 1.2 | 15.8 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 1.2 | 4.6 | GO:0071817 | MMXD complex(GO:0071817) |
| 1.1 | 9.7 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 1.1 | 4.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 1.0 | 28.2 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 1.0 | 3.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 1.0 | 3.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 1.0 | 10.0 | GO:0005869 | dynactin complex(GO:0005869) |
| 1.0 | 30.8 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.9 | 5.6 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.9 | 14.0 | GO:0034709 | methylosome(GO:0034709) |
| 0.9 | 13.8 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.8 | 4.2 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.8 | 6.6 | GO:0070187 | telosome(GO:0070187) |
| 0.8 | 4.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.8 | 50.8 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.8 | 5.6 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.8 | 7.6 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.8 | 6.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.8 | 2.3 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.7 | 9.6 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.7 | 8.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.7 | 8.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.7 | 4.9 | GO:0090661 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.7 | 2.8 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.7 | 13.7 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.7 | 5.4 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.7 | 2.0 | GO:0055087 | Ski complex(GO:0055087) |
| 0.6 | 4.5 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.6 | 14.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.6 | 13.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.6 | 3.0 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.6 | 35.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.6 | 14.9 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.6 | 6.8 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.6 | 5.0 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.6 | 6.6 | GO:0071203 | WASH complex(GO:0071203) |
| 0.5 | 2.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.5 | 15.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.5 | 47.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.5 | 5.7 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.5 | 3.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.5 | 33.9 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.5 | 26.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.5 | 44.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.5 | 2.0 | GO:0070288 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.5 | 4.7 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.5 | 4.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.5 | 12.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.5 | 4.6 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.4 | 3.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.4 | 2.5 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.4 | 101.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.4 | 17.8 | GO:0030684 | preribosome(GO:0030684) |
| 0.4 | 67.8 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.4 | 2.4 | GO:0030891 | VCB complex(GO:0030891) |
| 0.4 | 4.0 | GO:0032039 | integrator complex(GO:0032039) |
| 0.4 | 4.6 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.4 | 5.7 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.4 | 44.2 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.4 | 5.2 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.4 | 3.6 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.4 | 1.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.4 | 2.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.3 | 3.7 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.3 | 1.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.3 | 1.6 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.3 | 16.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.3 | 1.2 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.3 | 17.3 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.3 | 19.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.3 | 4.3 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.2 | 11.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.2 | 10.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 1.5 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.2 | 1.8 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 1.0 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.2 | 2.9 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.2 | 0.6 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.2 | 78.8 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.2 | 8.9 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.2 | 4.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.2 | 7.4 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 0.7 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.1 | 1.3 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 1.8 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 2.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 2.0 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 2.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 2.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 4.8 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 2.7 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 1.9 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 5.7 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 14.8 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.1 | 11.8 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 7.4 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 3.4 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 8.5 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 4.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 2.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 26.5 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 1.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 2.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 6.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 2.2 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 1.3 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 2.9 | GO:0005840 | ribosome(GO:0005840) |
| 0.1 | 9.6 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.1 | 3.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 2.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.8 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 1.7 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.3 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 1.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.4 | GO:0036379 | myofilament(GO:0036379) |
| 0.0 | 1.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 99.3 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 16.5 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.3 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 11.5 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.0 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 16.0 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 0.3 | GO:0000791 | euchromatin(GO:0000791) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 31.4 | 94.3 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 10.8 | 54.0 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 9.5 | 47.7 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 8.7 | 43.6 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 8.0 | 111.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 7.4 | 22.2 | GO:0003896 | DNA primase activity(GO:0003896) |
| 7.0 | 35.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 6.6 | 19.7 | GO:0008988 | rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) |
| 5.6 | 16.7 | GO:0033989 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 5.5 | 16.4 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 5.2 | 31.4 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 5.0 | 60.6 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 5.0 | 115.2 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 4.7 | 18.9 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 4.1 | 52.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 3.7 | 33.7 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 3.6 | 17.8 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 3.5 | 21.0 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 3.5 | 27.8 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 3.4 | 30.4 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 3.3 | 13.2 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 2.8 | 8.4 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 2.8 | 19.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 2.6 | 15.9 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 2.5 | 15.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 2.5 | 22.2 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 2.4 | 14.4 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 2.4 | 14.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 2.4 | 28.4 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 2.2 | 6.7 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 2.2 | 20.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 2.2 | 6.7 | GO:0055100 | adiponectin binding(GO:0055100) |
| 2.1 | 8.5 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 2.0 | 8.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 1.9 | 13.6 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 1.9 | 19.2 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 1.9 | 15.3 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 1.8 | 54.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 1.8 | 10.7 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 1.7 | 10.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 1.7 | 89.3 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 1.6 | 8.2 | GO:1903135 | cupric ion binding(GO:1903135) |
| 1.6 | 6.5 | GO:0070326 | lipase binding(GO:0035473) very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 1.6 | 57.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 1.6 | 6.4 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 1.6 | 29.9 | GO:0051400 | BH domain binding(GO:0051400) |
| 1.6 | 7.8 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 1.5 | 9.0 | GO:0004793 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 1.4 | 5.7 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 1.4 | 58.0 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 1.4 | 11.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 1.4 | 4.2 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 1.3 | 4.0 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 1.3 | 19.5 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 1.3 | 34.8 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 1.3 | 7.6 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 1.2 | 9.8 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 1.2 | 7.4 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 1.2 | 35.5 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 1.2 | 6.0 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 1.1 | 3.4 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 1.1 | 41.6 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 1.1 | 15.7 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 1.0 | 3.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 1.0 | 5.1 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 1.0 | 5.1 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.9 | 49.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.9 | 20.1 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.9 | 7.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.9 | 10.7 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.9 | 9.7 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.8 | 21.9 | GO:0046961 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.8 | 7.6 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.8 | 20.8 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.8 | 14.3 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.8 | 3.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.7 | 28.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.7 | 34.9 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.7 | 4.9 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.7 | 13.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.7 | 2.8 | GO:0032357 | oxidized purine DNA binding(GO:0032357) |
| 0.7 | 4.8 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.7 | 13.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.6 | 6.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.6 | 14.2 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.6 | 5.8 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.6 | 8.3 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.6 | 6.9 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.6 | 4.2 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.6 | 21.2 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.6 | 18.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.6 | 8.8 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.6 | 9.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.6 | 4.0 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.6 | 2.3 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.5 | 12.0 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.5 | 19.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.5 | 42.1 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.5 | 6.4 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.5 | 13.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.5 | 3.5 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.5 | 2.0 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.5 | 3.3 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.5 | 51.1 | GO:0003774 | motor activity(GO:0003774) |
| 0.4 | 3.1 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.4 | 5.0 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.4 | 21.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.4 | 17.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.4 | 3.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.4 | 2.3 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.4 | 79.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.4 | 10.5 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.4 | 7.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.3 | 40.2 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.3 | 2.3 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.3 | 0.7 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.3 | 2.6 | GO:0015288 | porin activity(GO:0015288) |
| 0.3 | 1.3 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.3 | 13.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.3 | 5.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.3 | 2.2 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.3 | 2.4 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.3 | 1.8 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.3 | 4.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.3 | 14.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.3 | 1.9 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.3 | 3.5 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.3 | 3.9 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.3 | 21.3 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.2 | 24.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.2 | 2.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.2 | 20.9 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.2 | 13.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.2 | 2.7 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.2 | 4.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.2 | 8.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.2 | 1.3 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.2 | 2.5 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.2 | 1.9 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.2 | 0.6 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.2 | 0.7 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.2 | 7.0 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 5.3 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 5.6 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.2 | 2.6 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.2 | 1.4 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.2 | 5.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.2 | 58.1 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.2 | 1.7 | GO:0031386 | protein tag(GO:0031386) |
| 0.2 | 4.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 23.6 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.2 | 5.1 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.2 | 8.8 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 4.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 1.4 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 7.5 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.1 | 0.5 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 2.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 3.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 1.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 1.0 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 4.1 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.1 | 0.4 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.1 | 2.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 4.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.8 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.1 | 0.2 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.1 | 1.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 1.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 26.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.0 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 4.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 2.6 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 1.0 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 1.8 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 15.5 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 3.0 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 3.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 0.1 | GO:0015489 | polyamine transmembrane transporter activity(GO:0015203) putrescine transmembrane transporter activity(GO:0015489) |
| 0.1 | 4.7 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 0.6 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 2.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.7 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 1.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 2.1 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 0.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 3.4 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.6 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 1.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 51.4 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 1.5 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 3.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 5.5 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 3.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 2.4 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 4.5 | GO:0022832 | voltage-gated ion channel activity(GO:0005244) voltage-gated channel activity(GO:0022832) |
| 0.0 | 0.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.6 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 66.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 2.6 | 124.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 1.9 | 43.5 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 1.8 | 47.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 1.6 | 39.9 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 1.0 | 51.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.9 | 47.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.8 | 23.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.7 | 39.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.7 | 28.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.7 | 12.0 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.6 | 21.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.6 | 31.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.5 | 16.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.5 | 10.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.5 | 18.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.4 | 48.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.3 | 7.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.3 | 19.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.3 | 1.8 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.2 | 16.2 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.2 | 8.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.2 | 10.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.2 | 24.4 | PID P73PATHWAY | p73 transcription factor network |
| 0.2 | 13.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 4.2 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 4.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 12.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 2.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 0.4 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 5.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 2.9 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 3.9 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 4.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.1 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 4.7 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 2.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 1.9 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 2.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.4 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.6 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 131.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 3.1 | 85.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 3.0 | 6.0 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 2.6 | 73.3 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 2.6 | 54.8 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 2.0 | 47.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 1.8 | 23.7 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 1.7 | 13.9 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 1.7 | 161.6 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 1.5 | 108.0 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 1.4 | 46.3 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 1.3 | 20.0 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 1.2 | 18.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 1.1 | 100.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 1.1 | 43.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.9 | 13.2 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.9 | 44.1 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.9 | 16.7 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.9 | 14.8 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.9 | 58.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.9 | 11.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.9 | 13.9 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.9 | 16.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.8 | 13.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.8 | 16.8 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.8 | 6.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.8 | 27.7 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.8 | 5.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.8 | 15.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.8 | 9.6 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.8 | 8.5 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.8 | 21.9 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.8 | 63.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.7 | 27.8 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.7 | 19.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.7 | 15.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.6 | 18.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.6 | 1.9 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.6 | 1.3 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.6 | 15.2 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.6 | 15.2 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.6 | 20.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.6 | 9.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.6 | 8.7 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.6 | 18.3 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.5 | 26.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.5 | 4.7 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.5 | 16.4 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.4 | 8.1 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.3 | 10.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.3 | 30.6 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.3 | 11.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.3 | 14.1 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.3 | 20.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.3 | 8.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.3 | 4.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.3 | 1.5 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.3 | 7.1 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 5.0 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.2 | 3.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 26.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.2 | 3.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 8.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.2 | 5.0 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.2 | 9.0 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.2 | 3.7 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.2 | 10.9 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.2 | 5.7 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 22.9 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.1 | 4.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.7 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 4.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 3.6 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 1.0 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.1 | 3.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 1.4 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |
| 0.1 | 2.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 0.6 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.3 | REACTOME S PHASE | Genes involved in S Phase |
| 0.0 | 0.8 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 1.1 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.8 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 4.0 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.0 | 0.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |