Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
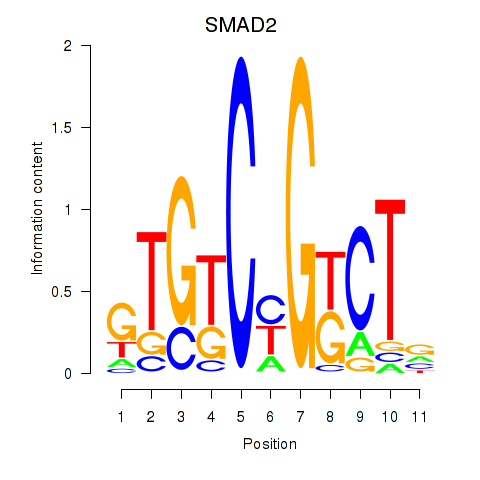
Results for SMAD2
Z-value: 3.96
Transcription factors associated with SMAD2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SMAD2
|
ENSG00000175387.16 | SMAD2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SMAD2 | hg38_v1_chr18_-_47930630_47930681 | -0.33 | 6.1e-07 | Click! |
Activity profile of SMAD2 motif
Sorted Z-values of SMAD2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SMAD2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_38561530 | 12.31 |
ENST00000378482.7
ENST00000286824.6 |
TSPAN7
|
tetraspanin 7 |
| chr1_+_160115715 | 10.42 |
ENST00000361216.8
|
ATP1A2
|
ATPase Na+/K+ transporting subunit alpha 2 |
| chr17_-_10114546 | 9.99 |
ENST00000323816.8
|
GAS7
|
growth arrest specific 7 |
| chr12_+_53050179 | 9.52 |
ENST00000546602.5
ENST00000552570.5 ENST00000549700.5 |
TNS2
|
tensin 2 |
| chr17_-_15262537 | 9.01 |
ENST00000395936.7
ENST00000675819.1 ENST00000674707.1 ENST00000675854.1 ENST00000426385.4 ENST00000395938.7 ENST00000612492.5 ENST00000675808.1 |
PMP22
|
peripheral myelin protein 22 |
| chr19_+_735026 | 8.76 |
ENST00000592155.5
ENST00000590161.2 |
PALM
|
paralemmin |
| chr5_-_151686908 | 8.46 |
ENST00000231061.9
|
SPARC
|
secreted protein acidic and cysteine rich |
| chr2_-_219308963 | 7.60 |
ENST00000423636.6
ENST00000442029.5 ENST00000412847.5 |
PTPRN
|
protein tyrosine phosphatase receptor type N |
| chr1_+_160127672 | 7.54 |
ENST00000447527.1
|
ATP1A2
|
ATPase Na+/K+ transporting subunit alpha 2 |
| chrX_-_107775740 | 7.25 |
ENST00000372383.9
|
TSC22D3
|
TSC22 domain family member 3 |
| chrX_-_107775951 | 6.72 |
ENST00000315660.8
ENST00000372384.6 ENST00000502650.1 ENST00000506724.1 |
TSC22D3
|
TSC22 domain family member 3 |
| chrX_+_102651366 | 6.72 |
ENST00000415986.5
ENST00000444152.5 ENST00000361600.9 |
GPRASP1
|
G protein-coupled receptor associated sorting protein 1 |
| chr5_+_141382702 | 6.06 |
ENST00000617050.1
ENST00000518325.2 |
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr1_+_2050387 | 5.72 |
ENST00000378567.8
|
PRKCZ
|
protein kinase C zeta |
| chr22_+_19718390 | 5.65 |
ENST00000383045.7
ENST00000438754.6 |
SEPTIN5
|
septin 5 |
| chr12_+_53050014 | 5.51 |
ENST00000314250.11
|
TNS2
|
tensin 2 |
| chr7_+_29194757 | 5.33 |
ENST00000222792.11
|
CHN2
|
chimerin 2 |
| chr19_+_35775515 | 4.95 |
ENST00000378944.9
|
ARHGAP33
|
Rho GTPase activating protein 33 |
| chr4_-_5888400 | 4.91 |
ENST00000397890.6
|
CRMP1
|
collapsin response mediator protein 1 |
| chr6_-_152168291 | 4.81 |
ENST00000354674.5
|
SYNE1
|
spectrin repeat containing nuclear envelope protein 1 |
| chr16_+_55479188 | 4.81 |
ENST00000219070.9
|
MMP2
|
matrix metallopeptidase 2 |
| chr6_-_152168349 | 4.77 |
ENST00000539504.5
|
SYNE1
|
spectrin repeat containing nuclear envelope protein 1 |
| chr11_-_117295485 | 4.67 |
ENST00000680971.1
|
BACE1
|
beta-secretase 1 |
| chr9_-_98708856 | 4.63 |
ENST00000259455.4
|
GABBR2
|
gamma-aminobutyric acid type B receptor subunit 2 |
| chr19_+_35775530 | 4.63 |
ENST00000314737.9
ENST00000007510.8 |
ARHGAP33
|
Rho GTPase activating protein 33 |
| chr6_+_39793008 | 4.57 |
ENST00000398904.6
|
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chr2_-_219309350 | 4.48 |
ENST00000295718.7
|
PTPRN
|
protein tyrosine phosphatase receptor type N |
| chr6_+_39792993 | 4.39 |
ENST00000538976.5
|
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chr12_-_55712402 | 4.39 |
ENST00000452168.6
|
ITGA7
|
integrin subunit alpha 7 |
| chr6_+_39792298 | 4.23 |
ENST00000633794.1
ENST00000274867.9 |
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chr12_-_6689244 | 4.07 |
ENST00000361959.7
ENST00000436774.6 ENST00000544482.1 |
ZNF384
|
zinc finger protein 384 |
| chr12_-_6689359 | 3.96 |
ENST00000683879.1
|
ZNF384
|
zinc finger protein 384 |
| chr22_+_22822658 | 3.91 |
ENST00000620395.2
|
IGLV2-8
|
immunoglobulin lambda variable 2-8 |
| chr9_-_133131109 | 3.85 |
ENST00000372062.8
|
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chr3_-_168095885 | 3.79 |
ENST00000470487.6
|
GOLIM4
|
golgi integral membrane protein 4 |
| chr16_+_90022600 | 3.77 |
ENST00000620723.4
ENST00000268699.9 |
GAS8
|
growth arrest specific 8 |
| chr11_-_66317037 | 3.77 |
ENST00000311330.4
|
CD248
|
CD248 molecule |
| chr19_+_49513154 | 3.73 |
ENST00000426395.7
ENST00000600273.5 ENST00000599988.5 |
FCGRT
|
Fc fragment of IgG receptor and transporter |
| chr19_+_11355386 | 3.67 |
ENST00000251473.9
ENST00000591329.5 ENST00000586380.5 |
PLPPR2
|
phospholipid phosphatase related 2 |
| chr22_+_22357739 | 3.67 |
ENST00000390294.2
|
IGLV1-47
|
immunoglobulin lambda variable 1-47 |
| chr22_+_22327298 | 3.67 |
ENST00000390291.2
|
IGLV1-50
|
immunoglobulin lambda variable 1-50 (non-functional) |
| chr19_-_14496088 | 3.58 |
ENST00000393033.9
ENST00000345425.6 ENST00000586027.5 ENST00000591349.5 ENST00000587210.1 |
GIPC1
|
GIPC PDZ domain containing family member 1 |
| chr1_-_206946448 | 3.50 |
ENST00000356495.5
|
PIGR
|
polymeric immunoglobulin receptor |
| chr3_+_124094663 | 3.47 |
ENST00000460856.5
ENST00000240874.7 |
KALRN
|
kalirin RhoGEF kinase |
| chr12_-_6689450 | 3.45 |
ENST00000355772.8
ENST00000417772.7 ENST00000319770.7 ENST00000396801.7 |
ZNF384
|
zinc finger protein 384 |
| chr2_-_219309484 | 3.42 |
ENST00000409251.7
ENST00000451506.5 ENST00000446182.5 |
PTPRN
|
protein tyrosine phosphatase receptor type N |
| chr19_-_14496144 | 3.38 |
ENST00000393028.5
|
GIPC1
|
GIPC PDZ domain containing family member 1 |
| chr7_-_102517755 | 3.37 |
ENST00000306682.6
ENST00000465829.6 ENST00000541662.5 |
RASA4B
|
RAS p21 protein activator 4B |
| chr9_+_15422704 | 3.37 |
ENST00000380821.7
ENST00000610884.4 ENST00000421710.5 |
SNAPC3
|
small nuclear RNA activating complex polypeptide 3 |
| chr7_-_102616692 | 3.20 |
ENST00000521076.5
ENST00000462172.5 ENST00000522801.5 ENST00000262940.12 ENST00000449970.6 |
RASA4
|
RAS p21 protein activator 4 |
| chr7_-_100896123 | 3.11 |
ENST00000428317.7
|
ACHE
|
acetylcholinesterase (Cartwright blood group) |
| chr1_+_159171607 | 3.11 |
ENST00000368124.8
ENST00000368125.9 ENST00000416746.1 |
CADM3
|
cell adhesion molecule 3 |
| chr22_-_20016807 | 3.08 |
ENST00000263207.8
|
ARVCF
|
ARVCF delta catenin family member |
| chr3_+_52495330 | 3.05 |
ENST00000321725.10
|
STAB1
|
stabilin 1 |
| chr13_-_96053370 | 3.03 |
ENST00000376712.4
ENST00000397618.7 ENST00000376747.8 ENST00000376714.7 ENST00000638479.1 ENST00000621375.5 |
UGGT2
|
UDP-glucose glycoprotein glucosyltransferase 2 |
| chr9_+_101028721 | 3.01 |
ENST00000374874.8
|
PLPPR1
|
phospholipid phosphatase related 1 |
| chr19_-_6481769 | 3.00 |
ENST00000381480.7
ENST00000543576.5 ENST00000590173.5 |
DENND1C
|
DENN domain containing 1C |
| chrX_-_63754664 | 3.00 |
ENST00000677315.1
ENST00000636392.1 ENST00000637040.1 ENST00000637178.1 ENST00000637557.1 ENST00000636048.1 ENST00000638021.1 ENST00000672513.1 |
ENSG00000288661.1
ARHGEF9
|
novel protein Cdc42 guanine nucleotide exchange factor 9 |
| chr11_-_33892010 | 2.84 |
ENST00000257818.3
|
LMO2
|
LIM domain only 2 |
| chr1_+_43946905 | 2.80 |
ENST00000372343.8
|
IPO13
|
importin 13 |
| chr13_-_26760741 | 2.74 |
ENST00000405846.5
|
GPR12
|
G protein-coupled receptor 12 |
| chr19_+_16888991 | 2.74 |
ENST00000248076.4
|
F2RL3
|
F2R like thrombin or trypsin receptor 3 |
| chr6_+_31615215 | 2.65 |
ENST00000337917.11
ENST00000376059.8 |
AIF1
|
allograft inflammatory factor 1 |
| chr17_+_3723889 | 2.59 |
ENST00000325418.5
|
HASPIN
|
histone H3 associated protein kinase |
| chr17_-_4142963 | 2.59 |
ENST00000381638.7
|
ZZEF1
|
zinc finger ZZ-type and EF-hand domain containing 1 |
| chr16_-_4801301 | 2.47 |
ENST00000586504.5
ENST00000649556.1 |
ROGDI
ENSG00000285952.1
|
rogdi atypical leucine zipper novel transcript |
| chr20_-_17682234 | 2.47 |
ENST00000377813.6
ENST00000377807.6 ENST00000360807.8 ENST00000398782.2 |
RRBP1
|
ribosome binding protein 1 |
| chr2_-_60550900 | 2.44 |
ENST00000643222.1
ENST00000643459.1 ENST00000489516.7 |
BCL11A
|
BAF chromatin remodeling complex subunit BCL11A |
| chrX_+_48801949 | 2.36 |
ENST00000376610.6
ENST00000462730.5 ENST00000376619.6 ENST00000465269.5 ENST00000334136.11 ENST00000476625.5 ENST00000646703.1 |
HDAC6
|
histone deacetylase 6 |
| chr14_+_100726883 | 2.36 |
ENST00000341267.9
ENST00000331224.10 ENST00000556051.1 |
DLK1
|
delta like non-canonical Notch ligand 1 |
| chr20_+_44715360 | 2.32 |
ENST00000190983.5
|
CCN5
|
cellular communication network factor 5 |
| chr16_+_2148603 | 2.28 |
ENST00000210187.11
|
RAB26
|
RAB26, member RAS oncogene family |
| chr1_-_201946571 | 2.26 |
ENST00000616739.1
|
LMOD1
|
leiomodin 1 |
| chr8_-_95269190 | 2.26 |
ENST00000286688.6
|
C8orf37
|
chromosome 8 open reading frame 37 |
| chr15_+_45587366 | 2.25 |
ENST00000220531.9
|
BLOC1S6
|
biogenesis of lysosomal organelles complex 1 subunit 6 |
| chr6_-_56954747 | 2.21 |
ENST00000680361.1
|
DST
|
dystonin |
| chr7_+_66629078 | 2.16 |
ENST00000449064.6
ENST00000638540.1 ENST00000640234.1 |
KCTD7
|
potassium channel tetramerization domain containing 7 |
| chr3_-_195876635 | 2.15 |
ENST00000672669.1
ENST00000672886.1 ENST00000672098.1 ENST00000671767.1 ENST00000672548.1 |
TNK2
|
tyrosine kinase non receptor 2 |
| chr13_-_26222255 | 2.14 |
ENST00000381588.9
|
RNF6
|
ring finger protein 6 |
| chr1_+_27726005 | 2.13 |
ENST00000530324.5
ENST00000234549.11 ENST00000373949.5 ENST00000010299.10 |
FAM76A
|
family with sequence similarity 76 member A |
| chr19_+_48469354 | 2.11 |
ENST00000452733.7
ENST00000641098.1 |
CYTH2
|
cytohesin 2 |
| chr7_+_66629023 | 2.06 |
ENST00000639879.1
ENST00000640851.1 |
KCTD7
|
potassium channel tetramerization domain containing 7 |
| chr16_+_28878480 | 2.06 |
ENST00000395503.9
|
ATP2A1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr1_+_36306359 | 2.06 |
ENST00000453908.8
|
SH3D21
|
SH3 domain containing 21 |
| chrX_+_153764178 | 2.02 |
ENST00000538966.5
|
PLXNB3
|
plexin B3 |
| chr11_+_14643826 | 2.02 |
ENST00000455098.2
|
PDE3B
|
phosphodiesterase 3B |
| chr22_-_38455199 | 2.02 |
ENST00000303592.3
|
KCNJ4
|
potassium inwardly rectifying channel subfamily J member 4 |
| chr5_-_74640575 | 2.00 |
ENST00000651128.1
|
ENC1
|
ectodermal-neural cortex 1 |
| chr1_+_27725945 | 1.95 |
ENST00000373954.11
ENST00000419687.6 |
FAM76A
|
family with sequence similarity 76 member A |
| chr21_-_37267511 | 1.93 |
ENST00000398998.1
|
VPS26C
|
VPS26 endosomal protein sorting factor C |
| chr15_+_45587580 | 1.92 |
ENST00000566801.5
ENST00000565323.6 ENST00000568816.5 |
BLOC1S6
|
biogenesis of lysosomal organelles complex 1 subunit 6 |
| chr6_+_101393699 | 1.91 |
ENST00000369134.9
ENST00000684068.1 ENST00000683903.1 ENST00000681975.1 |
GRIK2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr1_+_36307090 | 1.91 |
ENST00000505871.7
|
SH3D21
|
SH3 domain containing 21 |
| chr22_+_41560973 | 1.88 |
ENST00000306149.12
|
CSDC2
|
cold shock domain containing C2 |
| chrX_+_48802156 | 1.87 |
ENST00000643374.1
ENST00000644068.1 ENST00000441703.6 ENST00000643934.1 ENST00000489352.5 |
HDAC6
|
histone deacetylase 6 |
| chr1_-_151146643 | 1.87 |
ENST00000613223.1
|
SEMA6C
|
semaphorin 6C |
| chr1_-_151146611 | 1.84 |
ENST00000341697.7
ENST00000368914.8 |
SEMA6C
|
semaphorin 6C |
| chr12_-_10098977 | 1.82 |
ENST00000315330.8
ENST00000457018.6 |
CLEC1A
|
C-type lectin domain family 1 member A |
| chr22_-_24245059 | 1.81 |
ENST00000398292.3
ENST00000263112.11 ENST00000327365.10 ENST00000424217.1 |
GGT5
|
gamma-glutamyltransferase 5 |
| chr2_+_27014746 | 1.80 |
ENST00000648289.1
ENST00000458529.5 ENST00000402218.1 |
MAPRE3
|
microtubule associated protein RP/EB family member 3 |
| chr1_-_201946469 | 1.74 |
ENST00000367288.5
|
LMOD1
|
leiomodin 1 |
| chr7_+_102363874 | 1.74 |
ENST00000496391.5
|
PRKRIP1
|
PRKR interacting protein 1 |
| chr16_-_4937064 | 1.72 |
ENST00000590782.6
ENST00000345988.7 |
PPL
|
periplakin |
| chr3_+_124094696 | 1.70 |
ENST00000360013.7
ENST00000684186.1 ENST00000684276.1 |
KALRN
|
kalirin RhoGEF kinase |
| chr7_-_14841267 | 1.68 |
ENST00000406247.7
ENST00000399322.7 |
DGKB
|
diacylglycerol kinase beta |
| chr2_-_219243577 | 1.65 |
ENST00000409640.5
|
GLB1L
|
galactosidase beta 1 like |
| chrX_+_153494970 | 1.65 |
ENST00000331595.9
ENST00000431891.1 |
BGN
|
biglycan |
| chr6_+_32164586 | 1.63 |
ENST00000333845.11
ENST00000395512.5 ENST00000432129.1 |
EGFL8
|
EGF like domain multiple 8 |
| chr20_-_23086316 | 1.59 |
ENST00000246006.5
|
CD93
|
CD93 molecule |
| chrX_-_100732100 | 1.55 |
ENST00000372981.1
ENST00000263033.9 |
SYTL4
|
synaptotagmin like 4 |
| chr1_-_44017296 | 1.54 |
ENST00000357730.6
ENST00000360584.6 ENST00000528803.1 |
SLC6A9
|
solute carrier family 6 member 9 |
| chr22_+_37639660 | 1.48 |
ENST00000649765.2
ENST00000451997.6 |
SH3BP1
ENSG00000285304.1
|
SH3 domain binding protein 1 novel protein |
| chr4_-_121952014 | 1.42 |
ENST00000379645.8
|
TRPC3
|
transient receptor potential cation channel subfamily C member 3 |
| chr1_-_234479131 | 1.42 |
ENST00000040877.2
|
TARBP1
|
TAR (HIV-1) RNA binding protein 1 |
| chr19_+_11355491 | 1.41 |
ENST00000591608.1
|
PLPPR2
|
phospholipid phosphatase related 2 |
| chr2_-_227164194 | 1.40 |
ENST00000396625.5
|
COL4A4
|
collagen type IV alpha 4 chain |
| chr8_-_20303955 | 1.39 |
ENST00000381569.5
|
LZTS1
|
leucine zipper tumor suppressor 1 |
| chrX_+_153764233 | 1.38 |
ENST00000361971.10
|
PLXNB3
|
plexin B3 |
| chr11_+_14643782 | 1.37 |
ENST00000282096.9
|
PDE3B
|
phosphodiesterase 3B |
| chr1_-_44031352 | 1.28 |
ENST00000372306.7
ENST00000475075.6 |
SLC6A9
|
solute carrier family 6 member 9 |
| chr12_-_57016517 | 1.26 |
ENST00000441881.5
ENST00000458521.7 |
TAC3
|
tachykinin precursor 3 |
| chr17_+_7705193 | 1.23 |
ENST00000226091.3
|
EFNB3
|
ephrin B3 |
| chr7_+_155458129 | 1.19 |
ENST00000297375.4
|
EN2
|
engrailed homeobox 2 |
| chr13_-_26221703 | 1.19 |
ENST00000381570.7
ENST00000346166.7 |
RNF6
|
ring finger protein 6 |
| chr5_-_151141631 | 1.18 |
ENST00000523714.5
ENST00000521749.5 |
ANXA6
|
annexin A6 |
| chr16_+_2033264 | 1.17 |
ENST00000565855.5
ENST00000566198.1 |
SLC9A3R2
|
SLC9A3 regulator 2 |
| chr12_-_12266769 | 1.17 |
ENST00000543091.1
|
LRP6
|
LDL receptor related protein 6 |
| chr20_+_45469745 | 1.13 |
ENST00000372676.8
ENST00000217425.9 ENST00000339946.7 |
WFDC2
|
WAP four-disulfide core domain 2 |
| chr12_+_3491189 | 1.07 |
ENST00000382622.4
|
PRMT8
|
protein arginine methyltransferase 8 |
| chr17_-_3668640 | 1.07 |
ENST00000611779.4
|
TAX1BP3
|
Tax1 binding protein 3 |
| chr7_-_100895878 | 1.06 |
ENST00000419336.6
ENST00000241069.11 ENST00000411582.4 ENST00000302913.8 |
ACHE
|
acetylcholinesterase (Cartwright blood group) |
| chr14_+_80955577 | 1.01 |
ENST00000642209.1
ENST00000298171.7 |
TSHR
|
thyroid stimulating hormone receptor |
| chr1_-_39639626 | 0.98 |
ENST00000372852.4
|
HEYL
|
hes related family bHLH transcription factor with YRPW motif like |
| chr9_-_128067310 | 0.95 |
ENST00000373078.5
|
NAIF1
|
nuclear apoptosis inducing factor 1 |
| chr3_+_40505992 | 0.95 |
ENST00000420891.5
ENST00000314529.10 ENST00000418905.1 |
ZNF620
|
zinc finger protein 620 |
| chr18_+_13218195 | 0.92 |
ENST00000679167.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr17_-_3668557 | 0.92 |
ENST00000225525.4
|
TAX1BP3
|
Tax1 binding protein 3 |
| chr22_+_24495242 | 0.92 |
ENST00000382760.2
ENST00000326010.10 |
UPB1
|
beta-ureidopropionase 1 |
| chr14_+_101964561 | 0.91 |
ENST00000643508.2
ENST00000680137.1 ENST00000644881.2 ENST00000645149.2 ENST00000681574.1 ENST00000360184.10 ENST00000679720.1 ENST00000645114.2 |
DYNC1H1
|
dynein cytoplasmic 1 heavy chain 1 |
| chr1_+_15456727 | 0.91 |
ENST00000359621.5
|
CELA2A
|
chymotrypsin like elastase 2A |
| chr2_+_167868948 | 0.89 |
ENST00000392690.3
|
B3GALT1
|
beta-1,3-galactosyltransferase 1 |
| chr5_+_134526100 | 0.88 |
ENST00000395003.5
|
JADE2
|
jade family PHD finger 2 |
| chr6_+_41042557 | 0.83 |
ENST00000373158.6
ENST00000470917.1 |
TSPO2
|
translocator protein 2 |
| chr11_-_126062782 | 0.82 |
ENST00000531738.6
|
CDON
|
cell adhesion associated, oncogene regulated |
| chr16_+_57668299 | 0.81 |
ENST00000333493.9
ENST00000450388.7 |
ADGRG3
|
adhesion G protein-coupled receptor G3 |
| chr16_+_28878382 | 0.79 |
ENST00000357084.7
|
ATP2A1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr9_+_122375286 | 0.78 |
ENST00000373698.7
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr9_-_134917872 | 0.75 |
ENST00000616356.4
ENST00000371806.4 |
FCN1
|
ficolin 1 |
| chr19_-_43781249 | 0.73 |
ENST00000615047.4
|
KCNN4
|
potassium calcium-activated channel subfamily N member 4 |
| chr12_-_52814106 | 0.71 |
ENST00000551956.2
|
KRT4
|
keratin 4 |
| chr12_+_49961864 | 0.65 |
ENST00000293599.7
|
AQP5
|
aquaporin 5 |
| chr7_+_130380339 | 0.65 |
ENST00000481342.5
ENST00000604896.5 ENST00000011292.8 |
CPA1
|
carboxypeptidase A1 |
| chr19_-_40882226 | 0.63 |
ENST00000301146.9
|
CYP2A7
|
cytochrome P450 family 2 subfamily A member 7 |
| chr19_-_51646800 | 0.60 |
ENST00000599649.5
ENST00000429354.3 ENST00000360844.6 |
SIGLEC5
SIGLEC14
|
sialic acid binding Ig like lectin 5 sialic acid binding Ig like lectin 14 |
| chr4_+_92303946 | 0.59 |
ENST00000282020.9
|
GRID2
|
glutamate ionotropic receptor delta type subunit 2 |
| chr14_+_22226711 | 0.56 |
ENST00000390463.3
|
TRAV36DV7
|
T cell receptor alpha variable 36/delta variable 7 |
| chr1_+_37474572 | 0.55 |
ENST00000373087.7
|
ZC3H12A
|
zinc finger CCCH-type containing 12A |
| chr20_+_1895365 | 0.54 |
ENST00000358771.5
|
SIRPA
|
signal regulatory protein alpha |
| chr2_-_73284431 | 0.54 |
ENST00000521871.5
ENST00000520530.3 |
FBXO41
|
F-box protein 41 |
| chrX_-_30577759 | 0.53 |
ENST00000378962.4
|
TASL
|
TLR adaptor interacting with endolysosomal SLC15A4 |
| chr5_-_88731827 | 0.49 |
ENST00000627170.2
|
MEF2C
|
myocyte enhancer factor 2C |
| chr19_+_48624952 | 0.49 |
ENST00000599748.5
ENST00000599029.2 |
SPHK2
|
sphingosine kinase 2 |
| chr14_+_80955366 | 0.47 |
ENST00000342443.10
|
TSHR
|
thyroid stimulating hormone receptor |
| chr19_+_35936360 | 0.45 |
ENST00000246529.4
|
LRFN3
|
leucine rich repeat and fibronectin type III domain containing 3 |
| chrX_-_20141810 | 0.45 |
ENST00000379593.1
ENST00000379607.10 |
EIF1AX
|
eukaryotic translation initiation factor 1A X-linked |
| chr10_+_71212524 | 0.44 |
ENST00000335350.10
|
UNC5B
|
unc-5 netrin receptor B |
| chr10_-_124093582 | 0.44 |
ENST00000462406.1
ENST00000435907.6 |
CHST15
|
carbohydrate sulfotransferase 15 |
| chr1_+_16367088 | 0.42 |
ENST00000471507.5
ENST00000401089.3 ENST00000401088.9 ENST00000492354.1 |
SZRD1
|
SUZ RNA binding domain containing 1 |
| chr17_+_42288429 | 0.41 |
ENST00000676631.1
ENST00000677893.1 |
STAT5A
|
signal transducer and activator of transcription 5A |
| chr15_-_73368951 | 0.39 |
ENST00000261917.4
|
HCN4
|
hyperpolarization activated cyclic nucleotide gated potassium channel 4 |
| chr17_-_42185452 | 0.38 |
ENST00000293330.1
|
HCRT
|
hypocretin neuropeptide precursor |
| chr19_-_51630401 | 0.38 |
ENST00000683636.1
|
SIGLEC5
|
sialic acid binding Ig like lectin 5 |
| chr19_-_14114156 | 0.36 |
ENST00000589994.6
|
PRKACA
|
protein kinase cAMP-activated catalytic subunit alpha |
| chr1_+_2476284 | 0.34 |
ENST00000378486.8
|
PLCH2
|
phospholipase C eta 2 |
| chr4_-_41214535 | 0.33 |
ENST00000508593.6
|
APBB2
|
amyloid beta precursor protein binding family B member 2 |
| chr1_+_26159071 | 0.31 |
ENST00000374268.5
|
FAM110D
|
family with sequence similarity 110 member D |
| chr9_+_79572715 | 0.31 |
ENST00000265284.10
|
TLE4
|
TLE family member 4, transcriptional corepressor |
| chr22_+_37560472 | 0.25 |
ENST00000430687.1
ENST00000249014.5 |
CDC42EP1
|
CDC42 effector protein 1 |
| chr11_-_107719657 | 0.24 |
ENST00000525934.1
ENST00000531293.1 |
SLN
|
sarcolipin |
| chr3_-_9792404 | 0.24 |
ENST00000301964.7
|
TADA3
|
transcriptional adaptor 3 |
| chr1_-_160070150 | 0.21 |
ENST00000644903.1
|
KCNJ10
|
potassium inwardly rectifying channel subfamily J member 10 |
| chr22_-_36028773 | 0.19 |
ENST00000438146.7
|
RBFOX2
|
RNA binding fox-1 homolog 2 |
| chr3_-_165196369 | 0.18 |
ENST00000475390.2
|
SLITRK3
|
SLIT and NTRK like family member 3 |
| chr12_-_10098940 | 0.16 |
ENST00000420265.2
|
CLEC1A
|
C-type lectin domain family 1 member A |
| chr9_+_79573162 | 0.12 |
ENST00000425506.5
|
TLE4
|
TLE family member 4, transcriptional corepressor |
| chr1_+_99646025 | 0.12 |
ENST00000263174.9
ENST00000605497.5 ENST00000615664.1 |
PALMD
|
palmdelphin |
| chr11_-_22625804 | 0.11 |
ENST00000327470.6
|
FANCF
|
FA complementation group F |
| chr5_+_134526176 | 0.11 |
ENST00000681820.1
ENST00000512386.6 ENST00000612830.2 |
JADE2
|
jade family PHD finger 2 |
| chr13_-_41961078 | 0.09 |
ENST00000379310.8
ENST00000281496.6 |
VWA8
|
von Willebrand factor A domain containing 8 |
| chr1_-_160070102 | 0.07 |
ENST00000638728.1
ENST00000637644.1 |
KCNJ10
|
potassium inwardly rectifying channel subfamily J member 10 |
| chr11_+_59755427 | 0.04 |
ENST00000529177.5
|
STX3
|
syntaxin 3 |
| chr20_+_37346128 | 0.02 |
ENST00000373578.7
|
SRC
|
SRC proto-oncogene, non-receptor tyrosine kinase |
| chr17_-_41424583 | 0.01 |
ENST00000225550.4
|
KRT37
|
keratin 37 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 18.0 | GO:0051941 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 5.2 | 15.5 | GO:1904692 | positive regulation of type B pancreatic cell proliferation(GO:1904692) |
| 1.7 | 14.0 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 1.4 | 4.2 | GO:0070845 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 1.4 | 4.2 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 1.3 | 8.8 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 1.2 | 3.7 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 1.2 | 8.5 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 1.2 | 3.5 | GO:0002415 | immunoglobulin transcytosis in epithelial cells(GO:0002414) immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 1.1 | 5.7 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 1.0 | 3.0 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 1.0 | 4.8 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 1.0 | 9.6 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.9 | 2.8 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.9 | 2.8 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.9 | 5.6 | GO:1904526 | regulation of microtubule binding(GO:1904526) |
| 0.7 | 4.0 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.7 | 3.3 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.6 | 2.4 | GO:1904799 | negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.6 | 5.2 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.5 | 2.6 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) |
| 0.5 | 15.0 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.4 | 9.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.4 | 1.2 | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.4 | 3.4 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.4 | 3.4 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.4 | 1.5 | GO:1904588 | cellular response to glycoprotein(GO:1904588) cellular response to thyrotropin-releasing hormone(GO:1905229) |
| 0.4 | 1.4 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 0.3 | 2.7 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.3 | 0.9 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.3 | 1.4 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.3 | 4.2 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.3 | 3.0 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.3 | 0.8 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.2 | 0.7 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.2 | 7.0 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.2 | 3.8 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.2 | 1.0 | GO:0072014 | proximal tubule development(GO:0072014) |
| 0.2 | 2.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.2 | 0.5 | GO:0010587 | miRNA catabolic process(GO:0010587) negative regulation by host of viral genome replication(GO:0044828) |
| 0.2 | 0.9 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.2 | 2.0 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.2 | 0.8 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.2 | 1.9 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.3 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.1 | 1.3 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 1.8 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 1.2 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.1 | 7.2 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 2.8 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.1 | 0.5 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.1 | 6.7 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.1 | 0.7 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.1 | 0.8 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 | 4.6 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.1 | 1.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 13.1 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.1 | 5.4 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 0.9 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.6 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) positive regulation of long term synaptic depression(GO:1900454) |
| 0.1 | 1.7 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 1.4 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 4.7 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.1 | 1.2 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 4.4 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.1 | 0.2 | GO:1901876 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.1 | 2.0 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 2.1 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.4 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.4 | GO:0046449 | creatinine metabolic process(GO:0046449) |
| 0.1 | 2.1 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.9 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 9.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 7.6 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 3.4 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 1.2 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 1.6 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 1.4 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 18.2 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 2.3 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.4 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.0 | 0.8 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 3.0 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.4 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 | 3.3 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.6 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 2.5 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 0.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) definitive erythrocyte differentiation(GO:0060318) |
| 0.0 | 1.6 | GO:0042116 | macrophage activation(GO:0042116) |
| 0.0 | 0.3 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.2 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 3.0 | GO:0007265 | Ras protein signal transduction(GO:0007265) |
| 0.0 | 0.9 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.0 | 4.2 | GO:0051260 | protein homooligomerization(GO:0051260) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 1.2 | 4.6 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 1.0 | 18.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.7 | 9.6 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.7 | 5.7 | GO:0045179 | apical cortex(GO:0045179) |
| 0.4 | 2.2 | GO:0031673 | H zone(GO:0031673) |
| 0.4 | 8.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.4 | 6.6 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.3 | 2.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.3 | 4.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.3 | 10.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.3 | 4.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.3 | 9.0 | GO:0043218 | compact myelin(GO:0043218) |
| 0.2 | 3.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 1.4 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 25.1 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 0.8 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 2.8 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 2.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.5 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 5.2 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 3.0 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 4.4 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 4.0 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 1.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 3.8 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.1 | 1.9 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 2.6 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 3.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 1.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 3.5 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 3.1 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.2 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 4.9 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.6 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 2.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 3.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 9.6 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 3.9 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 3.0 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 5.8 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 3.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 3.0 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.0 | 0.8 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 2.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 3.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 8.6 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 6.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 3.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.8 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 24.7 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 5.9 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 0.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 3.4 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 14.0 | GO:0043426 | MRF binding(GO:0043426) |
| 1.4 | 18.0 | GO:1990239 | sodium:potassium-exchanging ATPase activity(GO:0005391) steroid hormone binding(GO:1990239) |
| 1.2 | 4.7 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 1.2 | 4.6 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 1.1 | 3.4 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 1.0 | 4.2 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.9 | 3.7 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.9 | 3.5 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.8 | 4.2 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.7 | 2.8 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.5 | 8.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.5 | 3.4 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.5 | 1.4 | GO:0070039 | rRNA (guanosine-2'-O-)-methyltransferase activity(GO:0070039) |
| 0.5 | 2.7 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.4 | 1.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.3 | 9.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.3 | 3.0 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.3 | 1.1 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.2 | 15.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 5.7 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.2 | 1.0 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.2 | 8.7 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.2 | 1.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 4.9 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 1.9 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 1.5 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.2 | 1.7 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.2 | 2.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.2 | 13.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.2 | 5.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.2 | 4.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 0.8 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.2 | 4.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 1.8 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 2.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.4 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.1 | 1.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 1.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 2.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 1.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 2.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.8 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 2.7 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.1 | 0.4 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.1 | 7.0 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 0.7 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 3.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.5 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 6.7 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.1 | 2.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 15.0 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.1 | 23.8 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.1 | 0.9 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.1 | 0.9 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 2.8 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 3.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 3.3 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 13.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.7 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 1.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 2.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.6 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.6 | GO:0005234 | ionotropic glutamate receptor activity(GO:0004970) extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 3.2 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 7.0 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 4.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.9 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.9 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 1.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 1.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 5.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.2 | 2.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 7.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 4.4 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 3.6 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 9.0 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 2.1 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 5.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 3.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 4.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 1.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.8 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 3.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 7.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 6.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.1 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 3.5 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 2.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.5 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.2 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 16.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 4.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 3.0 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.2 | 4.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 9.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.2 | 4.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 4.9 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 4.8 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.2 | 4.1 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 3.9 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 3.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 2.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 8.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.5 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 2.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 2.8 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 2.7 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 4.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 3.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.9 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 5.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 2.1 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 0.9 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 8.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 0.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 4.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |