Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
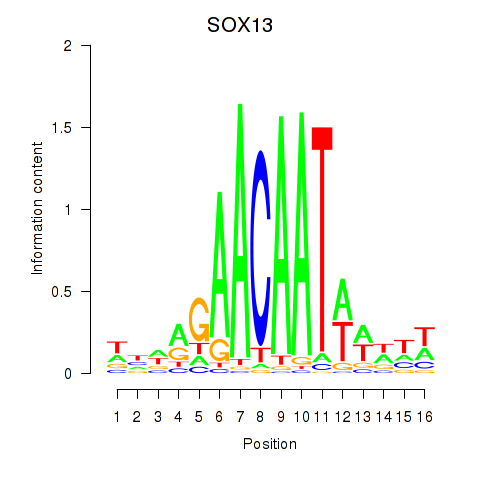
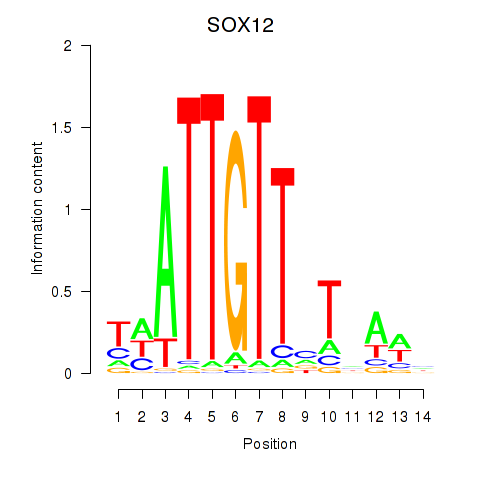
Results for SOX13_SOX12
Z-value: 3.87
Transcription factors associated with SOX13_SOX12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX13
|
ENSG00000143842.15 | SOX13 |
|
SOX12
|
ENSG00000177732.9 | SOX12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX13 | hg38_v1_chr1_+_204073104_204073121 | 0.17 | 1.3e-02 | Click! |
| SOX12 | hg38_v1_chr20_+_325536_325565 | -0.06 | 3.8e-01 | Click! |
Activity profile of SOX13_SOX12 motif
Sorted Z-values of SOX13_SOX12 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX13_SOX12
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 11.3 | GO:2000170 | positive regulation of actin filament-based movement(GO:1903116) positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 2.2 | 8.8 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 2.0 | 7.9 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 1.8 | 5.4 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 1.8 | 7.0 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 1.7 | 5.0 | GO:0034239 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 1.5 | 7.6 | GO:0035106 | operant conditioning(GO:0035106) |
| 1.5 | 6.0 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 1.4 | 28.9 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 1.3 | 3.9 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 1.2 | 3.5 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 1.1 | 4.5 | GO:1902081 | regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 1.1 | 3.2 | GO:0050720 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) interleukin-1 beta biosynthetic process(GO:0050720) |
| 1.1 | 4.3 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 1.1 | 3.2 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 1.0 | 2.9 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 1.0 | 3.9 | GO:0044782 | cilium organization(GO:0044782) |
| 0.9 | 6.2 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.9 | 5.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.8 | 3.3 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.8 | 10.7 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.8 | 8.4 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.7 | 3.7 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.7 | 7.9 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.7 | 2.9 | GO:0002005 | angiotensin catabolic process in blood(GO:0002005) |
| 0.7 | 5.0 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.7 | 2.1 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.7 | 2.8 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.7 | 2.1 | GO:0009720 | detection of hormone stimulus(GO:0009720) |
| 0.7 | 4.7 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.6 | 16.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.6 | 1.9 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.6 | 4.3 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.6 | 3.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.6 | 9.0 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.6 | 7.1 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.6 | 3.0 | GO:0010749 | regulation of nitric oxide mediated signal transduction(GO:0010749) negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.6 | 1.8 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.6 | 2.9 | GO:0003219 | cardiac right ventricle formation(GO:0003219) visceral serous pericardium development(GO:0061032) |
| 0.6 | 1.2 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.6 | 6.4 | GO:0035721 | intraciliary retrograde transport(GO:0035721) photoreceptor cell outer segment organization(GO:0035845) |
| 0.6 | 9.1 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.6 | 2.8 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.6 | 1.7 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.6 | 1.7 | GO:1990654 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 0.5 | 6.5 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.5 | 10.2 | GO:0021513 | spinal cord dorsal/ventral patterning(GO:0021513) |
| 0.5 | 3.2 | GO:1903282 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) regulation of peroxidase activity(GO:2000468) positive regulation of peroxidase activity(GO:2000470) |
| 0.5 | 1.6 | GO:0070340 | detection of bacterial lipoprotein(GO:0042494) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.5 | 4.2 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.5 | 4.6 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.5 | 50.9 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.5 | 3.0 | GO:1904304 | regulation of gastro-intestinal system smooth muscle contraction(GO:1904304) positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.5 | 10.5 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.5 | 2.0 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.5 | 1.0 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.5 | 4.4 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.5 | 1.9 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.5 | 1.4 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.5 | 1.9 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.5 | 3.3 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.5 | 1.4 | GO:0072757 | cellular response to camptothecin(GO:0072757) |
| 0.5 | 1.8 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.4 | 3.5 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.4 | 1.3 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.4 | 1.3 | GO:0046666 | retinal cell programmed cell death(GO:0046666) |
| 0.4 | 3.9 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.4 | 1.7 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.4 | 1.7 | GO:0070944 | neutrophil mediated cytotoxicity(GO:0070942) neutrophil mediated killing of symbiont cell(GO:0070943) neutrophil mediated killing of bacterium(GO:0070944) |
| 0.4 | 4.1 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.4 | 3.7 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.4 | 1.6 | GO:0050904 | diapedesis(GO:0050904) |
| 0.4 | 2.0 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.4 | 4.8 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.4 | 3.5 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.4 | 1.5 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.4 | 2.3 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.4 | 2.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.4 | 1.5 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.4 | 1.5 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.4 | 4.1 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.4 | 3.0 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.4 | 4.4 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.4 | 3.2 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.4 | 3.2 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.4 | 4.6 | GO:0006554 | lysine catabolic process(GO:0006554) |
| 0.3 | 2.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.3 | 1.0 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.3 | 14.2 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.3 | 4.0 | GO:0001787 | natural killer cell proliferation(GO:0001787) |
| 0.3 | 4.7 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.3 | 2.6 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.3 | 1.6 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.3 | 3.8 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.3 | 1.3 | GO:0036369 | traversing start control point of mitotic cell cycle(GO:0007089) transcription factor catabolic process(GO:0036369) cellular response to actinomycin D(GO:0072717) |
| 0.3 | 0.9 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.3 | 1.6 | GO:0014056 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) |
| 0.3 | 1.2 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.3 | 7.4 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.3 | 5.8 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.3 | 2.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.3 | 1.2 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.3 | 6.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.3 | 4.1 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.3 | 11.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.3 | 3.7 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.3 | 11.4 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.3 | 8.0 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.3 | 0.8 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.3 | 19.3 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.3 | 1.4 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.3 | 2.7 | GO:0045986 | negative regulation of smooth muscle contraction(GO:0045986) |
| 0.3 | 0.8 | GO:0060003 | copper ion export(GO:0060003) |
| 0.3 | 2.7 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.3 | 0.8 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.3 | 1.5 | GO:0060595 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) mammary gland specification(GO:0060594) fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) |
| 0.3 | 1.5 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.3 | 2.3 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 1.0 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.2 | 2.0 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.2 | 2.6 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.2 | 4.7 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.2 | 1.4 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.2 | 0.5 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.2 | 0.7 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.2 | 1.4 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.2 | 1.4 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.2 | 1.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.2 | 12.0 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.2 | 3.0 | GO:1901978 | positive regulation of cell cycle checkpoint(GO:1901978) |
| 0.2 | 1.4 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.2 | 2.0 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.2 | 2.0 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.2 | 1.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.2 | 1.1 | GO:1903936 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.2 | 2.0 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.2 | 1.5 | GO:1902172 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.2 | 4.1 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.2 | 0.6 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.2 | 1.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.2 | 2.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 0.6 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.2 | 1.0 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 | 0.4 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.2 | 2.2 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.2 | 0.4 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.2 | 3.6 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.2 | 9.8 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 1.7 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.2 | 2.9 | GO:1900115 | extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.2 | 7.2 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.2 | 22.5 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.2 | 3.1 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.2 | 2.5 | GO:0097503 | sialylation(GO:0097503) |
| 0.2 | 3.2 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.2 | 2.4 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.2 | 0.6 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.2 | 7.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 1.7 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.2 | 0.5 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.2 | 2.0 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.2 | 9.7 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 0.9 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 3.1 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.1 | 1.8 | GO:0001767 | establishment of lymphocyte polarity(GO:0001767) |
| 0.1 | 1.0 | GO:1903237 | regulation of leukocyte tethering or rolling(GO:1903236) negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.1 | 0.8 | GO:1903012 | positive regulation of bone development(GO:1903012) positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 0.5 | GO:1903629 | regulation of calcium-dependent ATPase activity(GO:1903610) negative regulation of calcium-dependent ATPase activity(GO:1903611) regulation of dUTP diphosphatase activity(GO:1903627) positive regulation of dUTP diphosphatase activity(GO:1903629) negative regulation of aminoacyl-tRNA ligase activity(GO:1903631) regulation of leucine-tRNA ligase activity(GO:1903633) negative regulation of leucine-tRNA ligase activity(GO:1903634) |
| 0.1 | 0.3 | GO:0032954 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.1 | 2.1 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 | 0.5 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 4.5 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.1 | 0.5 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 1.0 | GO:0060025 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) regulation of synaptic activity(GO:0060025) |
| 0.1 | 5.2 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 1.8 | GO:0006559 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 6.9 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.1 | 19.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.5 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.1 | 1.4 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.1 | 1.0 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 3.5 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.1 | 1.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.5 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.8 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.4 | GO:0071440 | regulation of histone H3-K14 acetylation(GO:0071440) positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 | 2.6 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 0.6 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.1 | 3.9 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 8.0 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 1.5 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 4.7 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.1 | 0.2 | GO:0046013 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 1.8 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.1 | 0.8 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 5.1 | GO:0032212 | positive regulation of telomere maintenance via telomerase(GO:0032212) |
| 0.1 | 3.3 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.1 | 9.6 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.1 | 14.7 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 1.9 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 14.6 | GO:0018210 | peptidyl-threonine phosphorylation(GO:0018107) peptidyl-threonine modification(GO:0018210) |
| 0.1 | 1.0 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.1 | 1.2 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.1 | 3.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 1.8 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.1 | 0.2 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.1 | 1.3 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 1.7 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.1 | 1.0 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.1 | 0.8 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.2 | GO:0043324 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.1 | 1.9 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 | 3.6 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 0.7 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 1.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 2.1 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.1 | 1.1 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.1 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 1.6 | GO:0045453 | bone resorption(GO:0045453) |
| 0.1 | 1.8 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.1 | 1.8 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.5 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 1.4 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.1 | 1.7 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 0.9 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.1 | 0.6 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.1 | 0.4 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 1.2 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 | 1.7 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 4.7 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 0.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 2.1 | GO:0051930 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.1 | 2.0 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.1 | 1.2 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.1 | 2.2 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 0.4 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.1 | 0.1 | GO:0035992 | tendon development(GO:0035989) tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 | 1.9 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.1 | 4.1 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 1.7 | GO:0042102 | positive regulation of T cell proliferation(GO:0042102) |
| 0.1 | 0.7 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 1.0 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.1 | 0.3 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.1 | 4.1 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 1.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.3 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.0 | 3.5 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 1.1 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.0 | 1.3 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.7 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 1.5 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.0 | 0.1 | GO:1903977 | positive regulation of glial cell migration(GO:1903977) |
| 0.0 | 2.8 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.4 | GO:0060547 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.0 | 0.9 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.0 | 2.9 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 2.8 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 2.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 1.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.6 | GO:0007130 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.0 | 0.6 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.0 | 0.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.6 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 6.3 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.2 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 4.0 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 0.2 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 2.4 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 0.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.3 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.6 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 2.9 | GO:0030856 | regulation of epithelial cell differentiation(GO:0030856) |
| 0.0 | 2.0 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.2 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 9.4 | GO:0048232 | spermatogenesis(GO:0007283) male gamete generation(GO:0048232) |
| 0.0 | 0.1 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 0.2 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 1.6 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.9 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 2.7 | GO:0051262 | protein tetramerization(GO:0051262) |
| 0.0 | 1.3 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.4 | GO:0010863 | positive regulation of phospholipase C activity(GO:0010863) |
| 0.0 | 2.1 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 1.2 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.1 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.3 | GO:0070206 | protein trimerization(GO:0070206) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 11.3 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 1.8 | 5.4 | GO:0098831 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) presynaptic cytoskeleton(GO:0099569) |
| 1.7 | 5.0 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 1.6 | 4.9 | GO:0033150 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) |
| 1.1 | 13.1 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 1.0 | 3.0 | GO:0044609 | DBIRD complex(GO:0044609) |
| 1.0 | 7.6 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.9 | 6.4 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.9 | 17.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.9 | 2.6 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.8 | 10.6 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.8 | 5.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.7 | 2.2 | GO:0031251 | PAN complex(GO:0031251) |
| 0.7 | 2.8 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.7 | 8.9 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.7 | 4.7 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.6 | 3.2 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.6 | 2.3 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.5 | 1.6 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.5 | 1.4 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.5 | 2.3 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.5 | 4.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.4 | 6.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.4 | 7.7 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.4 | 1.9 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.3 | 0.9 | GO:0033593 | BRCA2-MAGE-D1 complex(GO:0033593) |
| 0.3 | 0.9 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.3 | 9.4 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.3 | 2.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.3 | 1.7 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.3 | 2.0 | GO:1990745 | EARP complex(GO:1990745) |
| 0.3 | 1.7 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.3 | 13.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 11.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 8.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.2 | 2.1 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.2 | 2.7 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.2 | 9.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 0.9 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 19.6 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.2 | 4.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 1.7 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.2 | 0.6 | GO:0060987 | lipid tube(GO:0060987) |
| 0.2 | 3.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.2 | 3.3 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.2 | 12.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 3.5 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.2 | 1.7 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.2 | 2.5 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 3.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 1.0 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 1.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 1.9 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 5.0 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.7 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 3.9 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 0.6 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 1.6 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.1 | 2.1 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.1 | 4.1 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 1.4 | GO:0000800 | lateral element(GO:0000800) |
| 0.1 | 2.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 8.8 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 24.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 2.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 3.3 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 4.0 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 1.7 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 3.7 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 1.0 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 12.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.5 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 4.1 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 5.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 0.3 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 33.0 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 1.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 1.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 3.7 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 6.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 4.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 2.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 1.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 2.0 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 1.8 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 1.5 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 2.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 5.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 6.9 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.1 | 0.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 1.0 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 1.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 1.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 1.4 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.1 | 1.4 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 62.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.7 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 3.2 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 6.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 3.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 2.9 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 2.5 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 4.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 2.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 4.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 2.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.9 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 2.8 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 1.0 | GO:0097610 | cell division site(GO:0032153) cleavage furrow(GO:0032154) cell division site part(GO:0032155) cell surface furrow(GO:0097610) |
| 0.0 | 0.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 1.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.1 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 74.4 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 1.7 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 1.4 | GO:0016605 | PML body(GO:0016605) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.6 | GO:0016497 | substance K receptor activity(GO:0016497) |
| 2.4 | 7.2 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 2.1 | 6.3 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 1.9 | 41.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 1.5 | 6.0 | GO:0035473 | lipase binding(GO:0035473) |
| 1.4 | 4.3 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 1.4 | 8.4 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 1.4 | 4.2 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 1.3 | 4.0 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 1.1 | 3.3 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 1.1 | 3.2 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 1.1 | 3.2 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 1.1 | 2.1 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 1.1 | 7.4 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 1.0 | 10.3 | GO:0005549 | odorant binding(GO:0005549) |
| 1.0 | 4.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 1.0 | 2.9 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.9 | 3.8 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.8 | 2.5 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.8 | 3.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.8 | 9.1 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.8 | 2.3 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.7 | 5.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.7 | 2.8 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.7 | 2.7 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.7 | 4.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.7 | 2.0 | GO:0004750 | ribulose-phosphate 3-epimerase activity(GO:0004750) |
| 0.7 | 3.9 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.6 | 6.5 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.6 | 9.0 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.6 | 10.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.6 | 4.3 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.6 | 6.3 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.5 | 3.2 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.5 | 1.6 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.5 | 2.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.5 | 2.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.5 | 12.4 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.5 | 7.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.5 | 3.6 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.5 | 2.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.5 | 6.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.5 | 13.5 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.5 | 2.9 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.5 | 1.9 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.5 | 5.1 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.5 | 1.8 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.5 | 2.3 | GO:0030109 | HLA-B specific inhibitory MHC class I receptor activity(GO:0030109) |
| 0.5 | 2.3 | GO:0008948 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) oxaloacetate decarboxylase activity(GO:0008948) |
| 0.5 | 8.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.4 | 40.6 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.4 | 1.7 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.4 | 1.3 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.4 | 34.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.4 | 6.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.4 | 3.7 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.4 | 10.6 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.4 | 5.9 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.4 | 1.6 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.4 | 8.1 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) TFIIB-class transcription factor binding(GO:0001093) |
| 0.4 | 2.3 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.4 | 1.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.4 | 1.4 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.4 | 5.0 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.3 | 2.0 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.3 | 7.0 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.3 | 4.7 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.3 | 4.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 1.0 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.3 | 1.6 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.3 | 1.8 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.3 | 1.5 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.3 | 2.4 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.3 | 8.0 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.3 | 2.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.3 | 0.9 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.3 | 3.2 | GO:0004568 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.3 | 2.6 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.3 | 0.9 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.3 | 20.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.3 | 2.8 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.3 | 1.1 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.3 | 1.7 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.3 | 0.8 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.3 | 1.6 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.3 | 5.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.3 | 53.8 | GO:0003823 | antigen binding(GO:0003823) |
| 0.3 | 2.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.3 | 1.3 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.2 | 3.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 1.6 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.2 | 19.5 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.2 | 1.8 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 4.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 0.7 | GO:0004339 | glucan 1,4-alpha-glucosidase activity(GO:0004339) |
| 0.2 | 1.3 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.2 | 4.7 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.2 | 1.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 1.5 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 0.9 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.2 | 19.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 3.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 14.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 5.0 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.2 | 1.7 | GO:0019841 | retinol binding(GO:0019841) |
| 0.2 | 2.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.2 | 0.8 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.2 | 3.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 1.0 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.2 | 6.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 1.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 0.5 | GO:0001861 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 0.2 | 0.6 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.2 | 5.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 3.2 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.2 | 0.6 | GO:0043273 | CTPase activity(GO:0043273) |
| 0.1 | 4.3 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 5.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 1.7 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 0.5 | GO:0004341 | gluconolactonase activity(GO:0004341) |
| 0.1 | 0.8 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.1 | 1.9 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 1.5 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 3.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 1.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 1.9 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 4.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.4 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 3.5 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 14.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.7 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 2.2 | GO:0015144 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.1 | 3.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 2.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.8 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 1.0 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 3.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 3.5 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.1 | 3.0 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 0.4 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.8 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.9 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.1 | 1.9 | GO:0001163 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 1.4 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 6.1 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 0.7 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 2.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 4.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 5.3 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 2.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 4.5 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 5.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 1.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 8.1 | GO:0020037 | heme binding(GO:0020037) |
| 0.1 | 1.7 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 4.6 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.1 | 0.7 | GO:0046906 | tetrapyrrole binding(GO:0046906) |
| 0.1 | 0.3 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 1.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.6 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 1.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.6 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 1.0 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 8.9 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 1.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 1.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 2.5 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 1.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 3.1 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.9 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.2 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.5 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.8 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 1.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 2.3 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.6 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 1.5 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 1.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.1 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 1.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 3.4 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.5 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 6.0 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 3.1 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 1.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 4.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 14.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 52.7 | GO:0003677 | DNA binding(GO:0003677) |
| 0.0 | 1.7 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.1 | GO:0022840 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 1.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 1.4 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.4 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 10.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.4 | 19.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.3 | 18.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 2.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 3.2 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.2 | 37.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 8.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 7.3 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 2.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 8.8 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 31.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.9 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 1.3 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 7.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 0.9 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 0.9 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 3.2 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.1 | 2.0 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 4.4 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 4.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 2.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 3.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 3.0 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 4.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 1.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 0.8 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 2.0 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 5.5 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 4.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 2.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 1.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 2.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 0.7 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 1.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 2.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 2.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 2.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.7 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 1.6 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 6.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 11.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 3.2 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.6 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.9 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.9 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.5 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 14.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.7 | 10.9 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.7 | 11.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.7 | 13.8 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.7 | 8.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.5 | 11.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.5 | 5.0 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.5 | 44.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.3 | 4.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.3 | 6.1 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.3 | 6.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.3 | 2.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.3 | 6.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.2 | 18.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 4.8 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.2 | 2.3 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.2 | 2.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 4.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 6.2 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.2 | 3.4 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.2 | 7.9 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 4.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.2 | 4.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 4.7 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.1 | 2.0 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 8.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 1.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 5.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 12.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 0.9 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.5 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.1 | 1.6 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 3.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.8 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 2.8 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 14.6 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.1 | 1.5 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.1 | 4.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.3 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 2.1 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 24.4 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.1 | 2.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 0.6 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 1.7 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.0 | 1.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.6 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 1.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 3.8 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.4 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 1.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 6.5 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.9 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.8 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 1.8 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.4 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 2.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.1 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.5 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |