Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
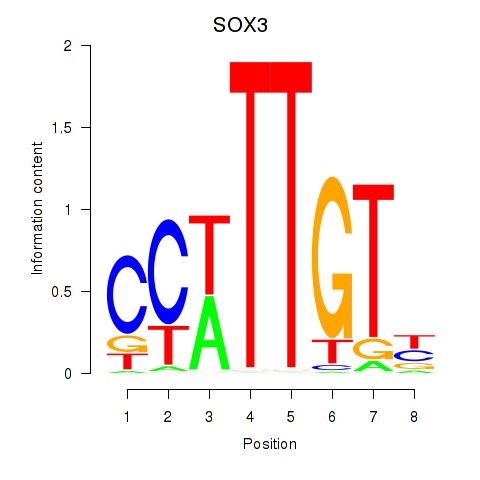
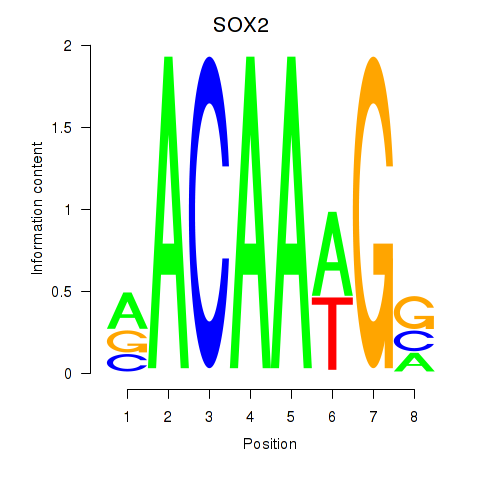
Results for SOX3_SOX2
Z-value: 0.64
Transcription factors associated with SOX3_SOX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX3
|
ENSG00000134595.9 | SOX3 |
|
SOX2
|
ENSG00000181449.4 | SOX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX2 | hg38_v1_chr3_+_181711915_181711934 | 0.33 | 8.0e-07 | Click! |
| SOX3 | hg38_v1_chrX_-_140505058_140505076 | 0.12 | 8.9e-02 | Click! |
Activity profile of SOX3_SOX2 motif
Sorted Z-values of SOX3_SOX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX3_SOX2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 25.2 | 125.8 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 15.9 | 47.8 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 13.5 | 53.9 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 13.0 | 39.0 | GO:1904616 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 12.6 | 50.3 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 11.9 | 143.1 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 9.2 | 55.4 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 8.4 | 25.1 | GO:1904397 | retinal rod cell differentiation(GO:0060221) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 6.0 | 83.5 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 5.9 | 5.9 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 5.6 | 39.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 5.6 | 27.8 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 5.3 | 21.2 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 4.3 | 55.4 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 4.2 | 70.9 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 4.0 | 20.1 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 3.9 | 34.9 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 3.8 | 11.5 | GO:2000830 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 3.8 | 22.9 | GO:0060313 | negative regulation of blood vessel remodeling(GO:0060313) |
| 3.6 | 36.4 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 3.6 | 18.0 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 3.5 | 10.6 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 3.4 | 17.1 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 3.4 | 13.6 | GO:0031179 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 3.3 | 20.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 3.3 | 92.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 3.3 | 9.9 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 3.3 | 9.8 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 3.2 | 9.7 | GO:0070256 | negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 3.2 | 51.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 3.1 | 15.7 | GO:0007412 | axon target recognition(GO:0007412) |
| 3.1 | 12.2 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 3.0 | 76.9 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 2.9 | 8.8 | GO:0061591 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 2.9 | 17.4 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 2.9 | 17.4 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 2.8 | 30.9 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 2.8 | 8.4 | GO:0030035 | microspike assembly(GO:0030035) |
| 2.7 | 24.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 2.7 | 5.4 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) |
| 2.7 | 8.0 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 2.7 | 8.0 | GO:1901388 | regulation of transforming growth factor beta activation(GO:1901388) negative regulation of transforming growth factor beta activation(GO:1901389) |
| 2.7 | 8.0 | GO:1904908 | negative regulation of maintenance of sister chromatid cohesion(GO:0034092) negative regulation of maintenance of mitotic sister chromatid cohesion(GO:0034183) maintenance of mitotic sister chromatid cohesion, telomeric(GO:0099403) mitotic sister chromatid cohesion, telomeric(GO:0099404) regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904907) negative regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904908) |
| 2.6 | 33.9 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 2.6 | 12.9 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 2.6 | 49.0 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 2.6 | 20.6 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 2.5 | 38.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 2.5 | 5.0 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 2.5 | 7.4 | GO:0060739 | mesenchymal-epithelial cell signaling involved in prostate gland development(GO:0060739) |
| 2.5 | 22.2 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 2.4 | 14.7 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 2.4 | 7.3 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 2.4 | 9.7 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 2.4 | 48.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 2.4 | 7.1 | GO:0008614 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 biosynthetic process(GO:0042819) |
| 2.3 | 14.0 | GO:0097338 | response to clozapine(GO:0097338) |
| 2.1 | 6.4 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 2.1 | 19.0 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 2.1 | 19.0 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) |
| 2.1 | 10.5 | GO:0021888 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 2.1 | 6.3 | GO:0042938 | dipeptide transport(GO:0042938) |
| 2.1 | 16.7 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 2.1 | 49.9 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 2.1 | 8.2 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 2.0 | 12.0 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 2.0 | 6.0 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 2.0 | 35.3 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 1.9 | 5.8 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 1.9 | 17.3 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 1.9 | 5.7 | GO:0070662 | mast cell proliferation(GO:0070662) |
| 1.9 | 9.3 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 1.8 | 7.3 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 1.8 | 16.2 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 1.8 | 10.7 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 1.8 | 10.6 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 1.7 | 5.2 | GO:0052026 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 1.7 | 22.6 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 1.7 | 5.2 | GO:0010768 | negative regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010768) transmission of virus(GO:0019089) dissemination or transmission of symbiont from host(GO:0044007) dissemination or transmission of organism from other organism involved in symbiotic interaction(GO:0051821) |
| 1.7 | 8.6 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 1.7 | 17.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 1.7 | 31.8 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 1.7 | 25.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 1.7 | 11.7 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 1.7 | 3.3 | GO:1904528 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) positive regulation of microtubule binding(GO:1904528) |
| 1.6 | 8.1 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 1.6 | 8.0 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 1.6 | 11.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 1.6 | 54.4 | GO:0048665 | neuron fate specification(GO:0048665) |
| 1.5 | 15.5 | GO:0070444 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 1.5 | 4.6 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 1.5 | 10.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 1.5 | 38.8 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 1.5 | 19.3 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 1.5 | 10.4 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 1.5 | 5.8 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 1.5 | 18.9 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 1.4 | 13.0 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 1.4 | 4.2 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 1.4 | 12.7 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 1.4 | 9.7 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 1.4 | 9.7 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 1.4 | 6.9 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 1.3 | 2.7 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 1.3 | 18.2 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 1.3 | 5.2 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 1.3 | 21.5 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 1.2 | 2.5 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 1.2 | 6.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 1.2 | 7.2 | GO:1903750 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 1.2 | 4.7 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 1.2 | 17.7 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 1.2 | 1.2 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 1.2 | 2.3 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 1.2 | 8.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 1.1 | 1.1 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 1.1 | 14.8 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 1.1 | 3.3 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 1.1 | 4.4 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 1.1 | 8.7 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 1.1 | 21.6 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 1.1 | 12.7 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 1.1 | 3.2 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 1.1 | 3.2 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 1.1 | 12.6 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 1.0 | 3.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 1.0 | 3.1 | GO:0061027 | umbilical cord morphogenesis(GO:0036304) umbilical cord development(GO:0061027) |
| 1.0 | 14.4 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 1.0 | 6.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 1.0 | 4.0 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 1.0 | 6.0 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 1.0 | 13.8 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 1.0 | 2.0 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 1.0 | 4.8 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 1.0 | 3.8 | GO:0006288 | base-excision repair, DNA ligation(GO:0006288) |
| 1.0 | 9.6 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 1.0 | 4.8 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.9 | 12.0 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) |
| 0.9 | 7.4 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.9 | 10.0 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.9 | 4.5 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.9 | 4.5 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.9 | 3.6 | GO:0035900 | response to isolation stress(GO:0035900) |
| 0.9 | 8.0 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.9 | 11.3 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.9 | 10.2 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.9 | 9.4 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.8 | 10.9 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.8 | 2.5 | GO:0007343 | egg activation(GO:0007343) |
| 0.8 | 10.8 | GO:2000790 | regulation of mesenchymal cell proliferation involved in lung development(GO:2000790) negative regulation of mesenchymal cell proliferation involved in lung development(GO:2000791) |
| 0.8 | 2.5 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.8 | 7.4 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.8 | 3.2 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 0.8 | 29.6 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.8 | 9.6 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.8 | 0.8 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.8 | 15.0 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.8 | 19.7 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.8 | 6.1 | GO:0060025 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) regulation of synaptic activity(GO:0060025) |
| 0.8 | 1.5 | GO:0043465 | fermentation(GO:0006113) regulation of fermentation(GO:0043465) |
| 0.8 | 133.6 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.8 | 27.2 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.8 | 3.8 | GO:0071287 | cellular response to manganese ion(GO:0071287) |
| 0.7 | 3.7 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.7 | 30.8 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.7 | 5.8 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.7 | 5.0 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.7 | 3.6 | GO:0022030 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.7 | 3.6 | GO:0033029 | regulation of neutrophil apoptotic process(GO:0033029) |
| 0.7 | 33.0 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.7 | 2.1 | GO:0099541 | trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) regulation of renin secretion into blood stream(GO:1900133) |
| 0.7 | 5.0 | GO:0030421 | defecation(GO:0030421) |
| 0.7 | 5.7 | GO:0021767 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.7 | 48.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.7 | 8.3 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.7 | 14.5 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.7 | 11.5 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.7 | 38.7 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.7 | 27.6 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.7 | 14.4 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.6 | 10.9 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.6 | 5.0 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.6 | 53.8 | GO:0030516 | regulation of axon extension(GO:0030516) |
| 0.6 | 2.5 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.6 | 6.1 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.6 | 3.0 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.6 | 4.2 | GO:2000507 | positive regulation of feeding behavior(GO:2000253) positive regulation of energy homeostasis(GO:2000507) |
| 0.6 | 4.1 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.6 | 1.7 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.6 | 1.2 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.6 | 5.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.5 | 3.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.5 | 2.7 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.5 | 2.7 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.5 | 4.3 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.5 | 10.8 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.5 | 3.2 | GO:0035604 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) fibroblast growth factor receptor signaling pathway involved in orbitofrontal cortex development(GO:0035607) |
| 0.5 | 3.2 | GO:1904261 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.5 | 1.0 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.5 | 81.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.5 | 2.5 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.5 | 4.9 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.5 | 8.7 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.5 | 6.7 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.5 | 11.8 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.5 | 1.4 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.5 | 1.4 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.5 | 10.2 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.5 | 9.6 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.5 | 0.5 | GO:0007227 | signal transduction downstream of smoothened(GO:0007227) |
| 0.4 | 9.3 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.4 | 3.9 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.4 | 6.6 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.4 | 11.7 | GO:0048675 | axon extension(GO:0048675) |
| 0.4 | 1.3 | GO:0033168 | positive regulation of Schwann cell differentiation(GO:0014040) conversion of ds siRNA to ss siRNA involved in RNA interference(GO:0033168) conversion of ds siRNA to ss siRNA(GO:0036404) |
| 0.4 | 1.7 | GO:0032599 | B cell receptor transport within lipid bilayer(GO:0032595) B cell receptor transport into membrane raft(GO:0032597) protein transport out of membrane raft(GO:0032599) chemokine receptor transport out of membrane raft(GO:0032600) negative regulation of transforming growth factor beta3 production(GO:0032913) chemokine receptor transport within lipid bilayer(GO:0033606) |
| 0.4 | 1.3 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.4 | 1.6 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.4 | 3.3 | GO:0009597 | detection of virus(GO:0009597) |
| 0.4 | 0.8 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.4 | 1.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.4 | 10.8 | GO:0006833 | water transport(GO:0006833) |
| 0.4 | 2.8 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.4 | 6.0 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.4 | 17.5 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.4 | 3.6 | GO:1903147 | negative regulation of macromitophagy(GO:1901525) negative regulation of mitophagy(GO:1903147) |
| 0.4 | 1.2 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.4 | 4.0 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.4 | 2.0 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.4 | 2.7 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.4 | 0.7 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.4 | 10.8 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.4 | 1.8 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.4 | 2.2 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.4 | 5.1 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.4 | 1.1 | GO:0010644 | cell communication by electrical coupling(GO:0010644) |
| 0.4 | 52.7 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.4 | 1.4 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.4 | 5.3 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.4 | 1.8 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.3 | 1.0 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.3 | 4.8 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.3 | 2.7 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.3 | 7.8 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.3 | 0.7 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.3 | 11.8 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.3 | 1.0 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.3 | 4.0 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.3 | 1.0 | GO:0015847 | putrescine transport(GO:0015847) |
| 0.3 | 4.3 | GO:1905024 | regulation of potassium ion export(GO:1902302) regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) regulation of potassium ion export across plasma membrane(GO:1903764) regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905024) regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905031) |
| 0.3 | 4.8 | GO:0010991 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.3 | 2.9 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.3 | 2.6 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.3 | 3.5 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.3 | 9.4 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.3 | 12.5 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.3 | 0.6 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.3 | 0.9 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.3 | 5.6 | GO:0043482 | pigment accumulation(GO:0043476) cellular pigment accumulation(GO:0043482) |
| 0.3 | 3.6 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.3 | 2.4 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.3 | 11.3 | GO:0001953 | negative regulation of cell-matrix adhesion(GO:0001953) |
| 0.3 | 12.1 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.3 | 1.5 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.3 | 1.5 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.3 | 31.8 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.3 | 1.1 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.3 | 2.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.3 | 2.5 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.3 | 3.3 | GO:0045779 | negative regulation of bone resorption(GO:0045779) |
| 0.3 | 1.6 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.3 | 1.1 | GO:0031645 | negative regulation of neurological system process(GO:0031645) |
| 0.3 | 6.4 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.3 | 7.6 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.2 | 7.4 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.2 | 4.7 | GO:0036109 | alpha-linolenic acid metabolic process(GO:0036109) |
| 0.2 | 1.5 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.2 | 0.7 | GO:0002215 | defense response to nematode(GO:0002215) regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.2 | 10.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.2 | 1.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.2 | 20.8 | GO:0001764 | neuron migration(GO:0001764) |
| 0.2 | 1.8 | GO:0015747 | urate transport(GO:0015747) |
| 0.2 | 1.4 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.2 | 5.8 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.2 | 2.4 | GO:0071073 | positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 0.2 | 0.6 | GO:0044771 | meiotic cell cycle checkpoint(GO:0033313) meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.2 | 3.0 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.2 | 5.3 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.2 | 0.8 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.2 | 0.6 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.2 | 1.8 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.2 | 0.8 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.2 | 3.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 2.5 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.2 | 1.0 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.2 | 1.3 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.2 | 3.5 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.2 | 5.2 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.2 | 0.4 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.2 | 4.2 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.2 | 1.8 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.2 | 0.5 | GO:0060613 | fat pad development(GO:0060613) |
| 0.2 | 12.1 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.2 | 3.6 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.2 | 0.3 | GO:0001300 | chronological cell aging(GO:0001300) positive regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905007) |
| 0.2 | 1.5 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.2 | 0.6 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.2 | 1.3 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.2 | 0.3 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.2 | 1.4 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.2 | 4.3 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) |
| 0.2 | 0.3 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.2 | 1.1 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.2 | 5.1 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.2 | 0.9 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 5.8 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.1 | 3.0 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.1 | 0.7 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.1 | 0.4 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.1 | 9.1 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 1.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 0.3 | GO:0048087 | positive regulation of developmental pigmentation(GO:0048087) |
| 0.1 | 7.7 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 1.2 | GO:0097012 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.1 | 2.1 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.1 | 0.8 | GO:1905146 | lysosomal protein catabolic process(GO:1905146) |
| 0.1 | 2.2 | GO:0050812 | regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.1 | 20.3 | GO:0007411 | axon guidance(GO:0007411) |
| 0.1 | 1.6 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.1 | 4.1 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.1 | 3.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 3.5 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.1 | 0.6 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 2.2 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 1.1 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 2.6 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.1 | 1.0 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 0.6 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) |
| 0.1 | 0.9 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 0.5 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 1.5 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.6 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.1 | 2.2 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 1.1 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.1 | 0.5 | GO:0015846 | polyamine transport(GO:0015846) |
| 0.1 | 4.4 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.1 | 0.8 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) |
| 0.1 | 2.7 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.1 | 1.7 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 0.1 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 4.3 | GO:0035308 | negative regulation of protein dephosphorylation(GO:0035308) |
| 0.1 | 2.0 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.7 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 1.0 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.1 | 0.3 | GO:0019477 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.1 | 0.3 | GO:2000314 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.1 | 1.4 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 3.8 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.1 | 1.0 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 1.4 | GO:0048662 | negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.1 | 0.7 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 1.9 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.1 | 0.8 | GO:0070193 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.1 | 1.2 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 4.2 | GO:0044070 | regulation of anion transport(GO:0044070) |
| 0.1 | 0.6 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.5 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 3.4 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.1 | 0.4 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 0.1 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 0.1 | 0.3 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 1.2 | GO:0014075 | response to amine(GO:0014075) |
| 0.0 | 1.1 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 1.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.7 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.8 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.6 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.2 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.0 | 0.3 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 2.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.3 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 3.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 3.2 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.6 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 | 0.7 | GO:0042993 | positive regulation of transcription factor import into nucleus(GO:0042993) |
| 0.0 | 5.2 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 4.6 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 2.4 | GO:0007626 | locomotory behavior(GO:0007626) |
| 0.0 | 1.0 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 1.4 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 1.0 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.3 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.0 | 0.2 | GO:1904350 | regulation of protein catabolic process in the vacuole(GO:1904350) positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.1 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.8 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.9 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.8 | GO:0001523 | retinoid metabolic process(GO:0001523) |
| 0.0 | 0.1 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.1 | GO:0021781 | glial cell fate commitment(GO:0021781) |
| 0.0 | 0.5 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.2 | GO:0034308 | primary alcohol metabolic process(GO:0034308) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.2 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 23.6 | 141.6 | GO:0033269 | internode region of axon(GO:0033269) |
| 5.1 | 20.3 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 4.9 | 14.6 | GO:0097229 | sperm end piece(GO:0097229) |
| 4.5 | 72.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 3.9 | 23.4 | GO:0045298 | tubulin complex(GO:0045298) |
| 3.8 | 38.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 3.6 | 64.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 3.3 | 20.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 3.1 | 15.5 | GO:0072534 | perineuronal net(GO:0072534) |
| 3.0 | 24.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 2.8 | 8.4 | GO:0044393 | microspike(GO:0044393) |
| 2.7 | 8.0 | GO:1990913 | sperm head plasma membrane(GO:1990913) ooplasm(GO:1990917) |
| 2.7 | 10.7 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 2.7 | 42.7 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 2.7 | 8.0 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 2.5 | 37.4 | GO:0097433 | dense body(GO:0097433) |
| 2.4 | 60.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 2.4 | 37.7 | GO:0042583 | chromaffin granule(GO:0042583) |
| 2.1 | 10.6 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 2.1 | 19.0 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 2.0 | 16.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 1.9 | 48.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 1.8 | 16.0 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 1.7 | 12.0 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 1.7 | 25.0 | GO:0030478 | actin cap(GO:0030478) |
| 1.6 | 121.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 1.5 | 4.6 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 1.4 | 55.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 1.4 | 13.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 1.4 | 25.9 | GO:0016342 | catenin complex(GO:0016342) |
| 1.3 | 9.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 1.2 | 3.6 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 1.1 | 8.8 | GO:0097386 | glial cell projection(GO:0097386) |
| 1.1 | 21.7 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 1.1 | 13.0 | GO:0032433 | filopodium tip(GO:0032433) |
| 1.0 | 3.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 1.0 | 8.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 1.0 | 177.1 | GO:0030426 | growth cone(GO:0030426) |
| 1.0 | 6.0 | GO:0032437 | cuticular plate(GO:0032437) |
| 1.0 | 103.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 1.0 | 29.6 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.9 | 405.2 | GO:0005874 | microtubule(GO:0005874) |
| 0.9 | 0.9 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.9 | 3.7 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.9 | 8.1 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.9 | 9.0 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.9 | 14.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.8 | 38.0 | GO:0031430 | M band(GO:0031430) |
| 0.8 | 3.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.8 | 3.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.8 | 162.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.7 | 191.4 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.7 | 119.3 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.7 | 12.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.7 | 1.5 | GO:0043203 | axon hillock(GO:0043203) |
| 0.7 | 13.6 | GO:0030904 | retromer complex(GO:0030904) |
| 0.7 | 8.9 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.7 | 62.3 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.7 | 14.7 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.7 | 14.0 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.6 | 2.5 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.6 | 5.5 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.6 | 4.8 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.6 | 17.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.6 | 1.2 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.6 | 20.2 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.5 | 10.9 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.5 | 11.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.5 | 1.5 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.5 | 103.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.5 | 1.4 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.5 | 23.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.5 | 5.0 | GO:0030673 | axolemma(GO:0030673) |
| 0.4 | 2.7 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.4 | 37.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.4 | 46.8 | GO:0030496 | midbody(GO:0030496) |
| 0.4 | 19.3 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.4 | 4.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.4 | 1.2 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.4 | 17.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.4 | 4.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.3 | 3.5 | GO:0071203 | WASH complex(GO:0071203) |
| 0.3 | 14.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.3 | 15.9 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.3 | 16.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.3 | 19.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.3 | 3.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.3 | 2.7 | GO:0032059 | bleb(GO:0032059) |
| 0.3 | 1.2 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.3 | 5.5 | GO:0005605 | basal lamina(GO:0005605) |
| 0.3 | 4.6 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.3 | 27.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.3 | 2.7 | GO:0010369 | chromocenter(GO:0010369) |
| 0.3 | 3.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.3 | 3.2 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.3 | 23.7 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.3 | 0.8 | GO:0071546 | pi-body(GO:0071546) |
| 0.3 | 0.8 | GO:0097637 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.3 | 6.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.2 | 6.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.2 | 2.6 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.2 | 7.8 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.2 | 16.6 | GO:0000922 | spindle pole(GO:0000922) |
| 0.2 | 3.9 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 10.0 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.2 | 2.4 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.2 | 1.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.2 | 1.4 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.2 | 0.8 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.2 | 3.5 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.2 | 3.2 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.2 | 0.8 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.2 | 0.8 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 11.3 | GO:0005819 | spindle(GO:0005819) |
| 0.1 | 0.3 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.1 | 23.6 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 0.4 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 1.7 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.3 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.1 | 0.3 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.1 | 8.2 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 2.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.3 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 3.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 0.6 | GO:0042589 | zymogen granule(GO:0042588) zymogen granule membrane(GO:0042589) |
| 0.1 | 7.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 17.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 3.9 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 1.5 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 5.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.3 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 2.2 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.7 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 1.2 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.9 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 2.9 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.1 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.0 | 1.0 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0033063 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.7 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 3.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.6 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.5 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 19.2 | 230.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 11.7 | 35.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 7.9 | 23.8 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 7.2 | 28.6 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 6.7 | 53.9 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 6.3 | 25.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 5.1 | 20.3 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 5.0 | 64.5 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 4.9 | 19.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 4.5 | 18.1 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 4.5 | 18.0 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 4.0 | 60.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 3.5 | 10.6 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 3.5 | 27.9 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 3.4 | 13.6 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 2.9 | 17.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 2.8 | 8.5 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 2.7 | 8.0 | GO:0002135 | CTP binding(GO:0002135) |
| 2.6 | 89.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 2.6 | 15.7 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 2.6 | 70.5 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 2.6 | 20.9 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 2.5 | 7.6 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 2.5 | 20.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 2.4 | 9.8 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 2.4 | 9.7 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 2.3 | 32.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 2.2 | 17.9 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 2.2 | 64.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 2.1 | 42.6 | GO:0031005 | filamin binding(GO:0031005) |
| 2.0 | 9.9 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 1.9 | 9.7 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 1.9 | 25.0 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 1.8 | 7.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 1.8 | 10.5 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 1.7 | 19.0 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 1.6 | 14.8 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 1.6 | 9.4 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 1.5 | 21.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 1.5 | 28.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 1.5 | 10.2 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 1.4 | 14.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 1.4 | 4.3 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 1.4 | 16.9 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 1.4 | 12.6 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 1.4 | 6.9 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 1.3 | 6.7 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 1.3 | 24.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 1.3 | 221.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 1.2 | 5.0 | GO:0071253 | connexin binding(GO:0071253) |
| 1.2 | 37.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 1.2 | 14.7 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 1.2 | 35.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 1.2 | 2.4 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 1.2 | 3.6 | GO:0005128 | erythropoietin receptor binding(GO:0005128) interleukin-3 receptor binding(GO:0005135) |
| 1.2 | 25.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 1.2 | 30.8 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 1.1 | 23.4 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 1.1 | 7.7 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 1.1 | 48.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 1.1 | 23.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 1.1 | 29.0 | GO:0070628 | proteasome binding(GO:0070628) |
| 1.1 | 3.2 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 1.1 | 17.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 1.1 | 17.0 | GO:0019841 | retinal binding(GO:0016918) retinol binding(GO:0019841) |
| 1.0 | 3.1 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 1.0 | 23.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 1.0 | 2.0 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 1.0 | 7.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 1.0 | 7.0 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 1.0 | 2.0 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 1.0 | 21.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 1.0 | 5.8 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 1.0 | 56.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 1.0 | 30.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.9 | 8.5 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.9 | 39.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.9 | 2.8 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.9 | 7.4 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.9 | 5.5 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.9 | 6.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.9 | 49.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.9 | 12.7 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.9 | 8.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.9 | 2.7 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.9 | 3.6 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.9 | 9.7 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.9 | 4.4 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.8 | 2.4 | GO:0070704 | C-5 sterol desaturase activity(GO:0000248) sterol desaturase activity(GO:0070704) |
| 0.8 | 11.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.8 | 2.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.8 | 3.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.8 | 3.8 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.7 | 10.8 | GO:0015250 | water channel activity(GO:0015250) |
| 0.7 | 249.8 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.7 | 2.8 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.7 | 2.0 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.7 | 19.7 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.7 | 11.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.6 | 3.2 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.6 | 9.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.6 | 3.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.6 | 2.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.6 | 3.6 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.6 | 0.6 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.6 | 2.3 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.6 | 14.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.5 | 1.6 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.5 | 11.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.5 | 2.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.5 | 52.6 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.5 | 5.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.5 | 3.6 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.5 | 1.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.5 | 4.6 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.5 | 2.5 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.5 | 2.9 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.5 | 2.4 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.5 | 1.4 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.5 | 2.4 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.5 | 5.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.5 | 8.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.5 | 6.0 | GO:0017136 | NAD-dependent histone deacetylase activity(GO:0017136) |
| 0.5 | 6.8 | GO:0042577 | phosphatidate phosphatase activity(GO:0008195) lipid phosphatase activity(GO:0042577) |
| 0.4 | 2.2 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.4 | 6.6 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.4 | 11.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.4 | 4.0 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.4 | 2.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.4 | 0.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.4 | 8.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.4 | 3.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.4 | 4.8 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.4 | 23.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.4 | 66.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.4 | 2.7 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.4 | 1.5 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.4 | 1.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.4 | 4.5 | GO:0036122 | BMP binding(GO:0036122) |
| 0.4 | 1.5 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.4 | 4.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.4 | 1.1 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.4 | 3.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.4 | 2.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.4 | 1.8 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.4 | 5.6 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.3 | 2.8 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.3 | 12.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.3 | 7.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.3 | 1.0 | GO:0033265 | choline binding(GO:0033265) |
| 0.3 | 21.8 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.3 | 1.4 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.3 | 6.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.3 | 1.0 | GO:0015489 | polyamine transmembrane transporter activity(GO:0015203) putrescine transmembrane transporter activity(GO:0015489) |
| 0.3 | 13.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.3 | 40.7 | GO:0005496 | steroid binding(GO:0005496) |
| 0.3 | 9.3 | GO:0001848 | complement binding(GO:0001848) |
| 0.3 | 2.7 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.3 | 0.6 | GO:0030613 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 0.3 | 3.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.3 | 15.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.3 | 13.8 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.3 | 1.5 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.3 | 2.9 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.3 | 1.2 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.3 | 6.4 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.3 | 1.4 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.3 | 4.0 | GO:0031432 | titin binding(GO:0031432) structural molecule activity conferring elasticity(GO:0097493) |
| 0.3 | 4.0 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.3 | 202.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.3 | 8.1 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.3 | 1.3 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.3 | 1.3 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.3 | 5.6 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.2 | 1.0 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.2 | 3.5 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.2 | 11.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 10.3 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.2 | 13.4 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.2 | 1.5 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.2 | 0.4 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.2 | 4.8 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.2 | 0.8 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.2 | 0.4 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.2 | 4.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 4.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.2 | 2.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.2 | 0.4 | GO:0046790 | virion binding(GO:0046790) |
| 0.2 | 42.0 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.2 | 1.8 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 1.0 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.2 | 65.8 | GO:0003779 | actin binding(GO:0003779) |
| 0.2 | 0.8 | GO:0047298 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.2 | 1.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.2 | 4.7 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.2 | 1.2 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.2 | 5.0 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.2 | 0.8 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.2 | 1.3 | GO:0015172 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.2 | 1.5 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.2 | 3.2 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.2 | 8.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 3.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 25.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.2 | 2.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 1.3 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 1.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 2.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 9.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.6 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 1.9 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.1 | 3.9 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 1.6 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 3.0 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 1.7 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 1.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 3.6 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 1.8 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 2.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 1.9 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.1 | 3.0 | GO:0050997 | quaternary ammonium group binding(GO:0050997) |
| 0.1 | 0.5 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 47.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 3.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 4.1 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 4.0 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 3.6 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 0.7 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.1 | 3.8 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 4.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.6 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.1 | 1.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.8 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.1 | 0.4 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 1.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 2.7 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 0.9 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.4 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 5.8 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 0.9 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 1.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 0.6 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 21.0 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 0.3 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.4 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 2.2 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.0 | 2.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.1 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.0 | 1.0 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 3.6 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.6 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 1.0 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 2.3 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.6 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 2.2 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.7 | 153.9 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 1.5 | 44.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 1.5 | 119.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 1.2 | 3.6 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 1.1 | 2.1 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 1.1 | 30.7 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.9 | 54.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.8 | 51.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.8 | 39.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.8 | 32.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.6 | 30.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.6 | 43.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.6 | 6.4 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.5 | 9.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.5 | 20.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.4 | 4.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.4 | 9.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.4 | 12.0 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.4 | 28.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.3 | 7.0 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.3 | 19.6 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.3 | 10.6 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.3 | 7.8 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.3 | 13.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.3 | 16.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.3 | 17.3 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.3 | 11.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.3 | 12.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.3 | 14.3 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.3 | 7.1 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.2 | 6.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.2 | 21.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.2 | 6.8 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.2 | 6.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.2 | 3.0 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.2 | 5.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.2 | 4.2 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.2 | 4.3 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 0.6 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 4.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 0.5 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 2.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 11.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 4.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 3.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 3.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 2.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 2.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 1.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 3.6 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 1.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.0 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 6.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.8 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 162.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 4.5 | 124.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 3.8 | 45.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 1.7 | 90.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 1.5 | 80.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 1.4 | 27.1 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 1.3 | 66.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 1.0 | 5.0 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.9 | 25.0 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.8 | 23.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.8 | 33.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.8 | 10.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.8 | 14.0 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.8 | 33.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.7 | 11.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.7 | 20.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.7 | 9.9 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.6 | 26.0 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.6 | 17.6 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.6 | 14.4 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.6 | 1.2 | REACTOME LAGGING STRAND SYNTHESIS | Genes involved in Lagging Strand Synthesis |
| 0.6 | 11.5 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.5 | 12.8 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.5 | 6.7 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.5 | 8.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.5 | 14.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.5 | 16.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.5 | 3.2 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.4 | 12.7 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.4 | 13.0 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.4 | 12.0 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.4 | 8.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.4 | 24.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.4 | 1.4 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.3 | 19.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.3 | 5.0 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.3 | 3.6 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.3 | 14.3 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.3 | 17.1 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.3 | 9.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.3 | 1.6 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.3 | 4.9 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.3 | 4.7 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 6.0 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.2 | 6.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.2 | 18.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.2 | 1.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 3.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.2 | 6.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 1.0 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 5.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 3.6 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.2 | 7.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.2 | 5.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 1.7 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.2 | 4.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 2.0 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.2 | 7.1 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 1.3 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 3.0 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 4.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 14.8 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 2.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 2.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.5 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 6.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 5.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 4.7 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.1 | 0.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.1 | 4.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 2.6 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 7.2 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 0.8 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 1.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.2 | REACTOME GABA B RECEPTOR ACTIVATION | Genes involved in GABA B receptor activation |
| 0.1 | 1.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 0.9 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 0.4 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 3.7 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.7 | REACTOME PROTEIN FOLDING | Genes involved in Protein folding |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.4 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.5 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |