Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
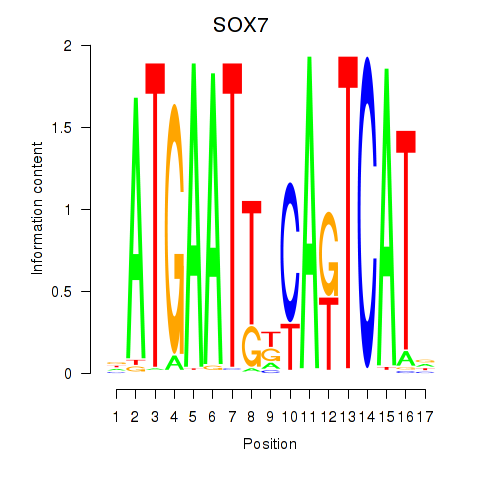
Results for SOX7
Z-value: 3.78
Transcription factors associated with SOX7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX7
|
ENSG00000171056.8 | SOX7 |
|
PINX1
|
ENSG00000258724.1 | PINX1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX7 | hg38_v1_chr8_-_10730498_10730525 | 0.62 | 1.3e-24 | Click! |
Activity profile of SOX7 motif
Sorted Z-values of SOX7 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX7
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_35241027 | 10.92 |
ENST00000330501.12
ENST00000601719.1 ENST00000591206.5 ENST00000261333.10 ENST00000355632.8 ENST00000585800.1 |
ZNF397
|
zinc finger protein 397 |
| chr16_-_33845229 | 9.34 |
ENST00000569103.2
|
IGHV3OR16-17
|
immunoglobulin heavy variable 3/OR16-17 (non-functional) |
| chr2_-_89100352 | 8.15 |
ENST00000479981.1
|
IGKV1-16
|
immunoglobulin kappa variable 1-16 |
| chr15_+_75347030 | 8.00 |
ENST00000566313.5
ENST00000355059.9 ENST00000568059.1 ENST00000568881.1 |
NEIL1
|
nei like DNA glycosylase 1 |
| chr14_-_106117159 | 7.76 |
ENST00000390601.3
|
IGHV3-11
|
immunoglobulin heavy variable 3-11 |
| chr17_-_35880350 | 6.58 |
ENST00000605140.6
ENST00000651122.1 ENST00000603197.6 |
CCL5
|
C-C motif chemokine ligand 5 |
| chr1_+_158931539 | 6.28 |
ENST00000368140.6
ENST00000368138.7 ENST00000392254.6 ENST00000392252.7 ENST00000368135.4 |
PYHIN1
|
pyrin and HIN domain family member 1 |
| chr16_+_33009175 | 6.23 |
ENST00000565407.2
|
IGHV3OR16-8
|
immunoglobulin heavy variable 3/OR16-8 (non-functional) |
| chr1_+_100133135 | 5.39 |
ENST00000370143.5
ENST00000370141.7 |
TRMT13
|
tRNA methyltransferase 13 homolog |
| chr8_+_28338640 | 5.33 |
ENST00000522209.1
|
PNOC
|
prepronociceptin |
| chr9_+_102995308 | 4.85 |
ENST00000612124.4
ENST00000374798.8 ENST00000487798.5 |
CYLC2
|
cylicin 2 |
| chr8_-_19602484 | 4.74 |
ENST00000454498.6
ENST00000520003.5 |
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr2_-_89222461 | 4.56 |
ENST00000482769.1
|
IGKV2-28
|
immunoglobulin kappa variable 2-28 |
| chr5_+_162067858 | 4.49 |
ENST00000361925.9
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr5_+_162067458 | 4.26 |
ENST00000639975.1
ENST00000639111.2 ENST00000639683.1 |
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr5_+_162067500 | 3.88 |
ENST00000639384.1
ENST00000640985.1 ENST00000638772.1 |
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr19_-_2783308 | 3.65 |
ENST00000677562.1
ENST00000677754.1 |
SGTA
|
small glutamine rich tetratricopeptide repeat containing alpha |
| chr19_-_2783241 | 3.64 |
ENST00000676943.1
ENST00000589251.5 ENST00000221566.7 ENST00000676984.1 |
SGTA
|
small glutamine rich tetratricopeptide repeat containing alpha |
| chr1_+_10450004 | 3.48 |
ENST00000377049.4
|
CORT
|
cortistatin |
| chr16_-_229398 | 3.36 |
ENST00000430864.5
ENST00000293872.13 ENST00000629543.2 ENST00000337351.8 ENST00000397783.5 |
LUC7L
|
LUC7 like |
| chr22_+_22720615 | 3.35 |
ENST00000390309.2
|
IGLV3-19
|
immunoglobulin lambda variable 3-19 |
| chr6_+_150368997 | 3.35 |
ENST00000392255.7
ENST00000500320.7 ENST00000344419.8 |
IYD
|
iodotyrosine deiodinase |
| chr5_+_162067764 | 3.24 |
ENST00000639213.2
ENST00000414552.6 |
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr13_-_37059432 | 3.22 |
ENST00000464744.5
|
SUPT20H
|
SPT20 homolog, SAGA complex component |
| chr2_+_3658193 | 3.16 |
ENST00000252505.4
|
ALLC
|
allantoicase |
| chr4_+_76074701 | 2.94 |
ENST00000355810.9
ENST00000349321.7 |
ART3
|
ADP-ribosyltransferase 3 (inactive) |
| chr6_+_131573219 | 2.87 |
ENST00000356962.2
ENST00000368087.8 ENST00000673427.1 ENST00000640973.1 |
ARG1
|
arginase 1 |
| chr4_-_47981535 | 2.58 |
ENST00000402813.9
|
CNGA1
|
cyclic nucleotide gated channel subunit alpha 1 |
| chr1_-_158426237 | 2.20 |
ENST00000641042.1
|
OR10K2
|
olfactory receptor family 10 subfamily K member 2 |
| chr10_+_94402486 | 2.18 |
ENST00000225235.5
|
TBC1D12
|
TBC1 domain family member 12 |
| chr6_+_42563981 | 2.02 |
ENST00000372899.6
ENST00000372901.2 |
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr6_+_150368892 | 1.88 |
ENST00000229447.9
ENST00000392256.6 |
IYD
|
iodotyrosine deiodinase |
| chr4_+_68447453 | 1.86 |
ENST00000305363.9
|
TMPRSS11E
|
transmembrane serine protease 11E |
| chr8_+_12945667 | 1.85 |
ENST00000524591.7
|
TRMT9B
|
tRNA methyltransferase 9B (putative) |
| chr21_-_14546297 | 1.54 |
ENST00000400566.6
ENST00000400564.5 |
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr1_+_200027702 | 1.35 |
ENST00000367362.8
|
NR5A2
|
nuclear receptor subfamily 5 group A member 2 |
| chr16_-_20352707 | 1.15 |
ENST00000396134.6
ENST00000573567.5 ENST00000570757.5 ENST00000396138.9 ENST00000571174.5 ENST00000576688.2 |
UMOD
|
uromodulin |
| chr4_-_103719980 | 0.95 |
ENST00000304883.3
|
TACR3
|
tachykinin receptor 3 |
| chr7_-_138755892 | 0.73 |
ENST00000644341.1
ENST00000478480.2 |
ATP6V0A4
|
ATPase H+ transporting V0 subunit a4 |
| chr10_+_4963406 | 0.66 |
ENST00000380872.9
ENST00000442997.5 |
AKR1C1
|
aldo-keto reductase family 1 member C1 |
| chr1_+_86993009 | 0.65 |
ENST00000370548.3
|
ENSG00000267561.2
|
novel protein |
| chr1_+_200027605 | 0.63 |
ENST00000236914.7
|
NR5A2
|
nuclear receptor subfamily 5 group A member 2 |
| chr11_-_62707413 | 0.51 |
ENST00000360796.10
ENST00000449636.6 |
BSCL2
|
BSCL2 lipid droplet biogenesis associated, seipin |
| chr11_+_7597182 | 0.46 |
ENST00000528883.5
|
PPFIBP2
|
PPFIA binding protein 2 |
| chr12_-_57050102 | 0.31 |
ENST00000300119.8
|
MYO1A
|
myosin IA |
| chr8_+_127409026 | 0.26 |
ENST00000465342.4
|
POU5F1B
|
POU class 5 homeobox 1B |
| chr12_-_10420550 | 0.15 |
ENST00000381903.2
ENST00000396439.7 |
KLRC3
|
killer cell lectin like receptor C3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.3 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 1.6 | 6.6 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 1.6 | 4.7 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 1.1 | 3.2 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.7 | 8.0 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.7 | 2.9 | GO:0043091 | L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 0.6 | 15.9 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.4 | 2.0 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.4 | 5.2 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.4 | 2.0 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.4 | 1.2 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.3 | 3.5 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.2 | 5.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.2 | 29.3 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 0.7 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.1 | 0.9 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 3.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 2.9 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 2.6 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.1 | 2.2 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 0.7 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 5.3 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 1.5 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.0 | 0.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 1.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 3.2 | GO:0007369 | gastrulation(GO:0007369) |
| 0.0 | 0.3 | GO:0030033 | microvillus assembly(GO:0030033) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0033011 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) |
| 1.2 | 3.5 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.4 | 15.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 3.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.2 | 3.4 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 2.6 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 4.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 6.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.7 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 2.9 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 5.3 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 2.2 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 3.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 6.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 18.9 | GO:0015630 | microtubule cytoskeleton(GO:0015630) |
| 0.0 | 2.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.6 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 1.0 | 5.2 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 1.0 | 7.3 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 1.0 | 15.9 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.9 | 4.7 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.8 | 5.3 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.6 | 2.9 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.4 | 8.0 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.4 | 3.4 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.4 | 6.0 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.3 | 2.0 | GO:0070728 | leucine binding(GO:0070728) |
| 0.2 | 0.7 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.2 | 5.4 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.2 | 2.6 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.2 | 0.9 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 30.1 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 3.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 1.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.7 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 1.5 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 4.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 2.0 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 1.6 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 2.2 | GO:0017137 | Rab GTPase binding(GO:0017137) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 2.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 2.9 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 15.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 5.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 4.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 6.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 6.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 2.0 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.7 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 2.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 2.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |