Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for SPIC
Z-value: 4.77
Transcription factors associated with SPIC
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SPIC
|
ENSG00000166211.8 | SPIC |
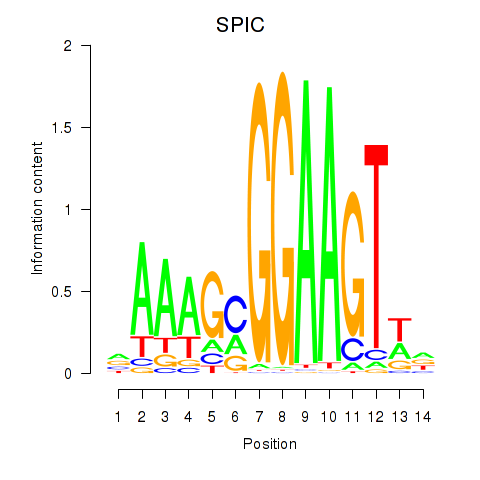
Activity profile of SPIC motif
Sorted Z-values of SPIC motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SPIC
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 29.0 | 86.9 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 23.1 | 69.2 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 21.1 | 105.6 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 18.2 | 72.7 | GO:2001193 | gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) |
| 16.5 | 49.6 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 15.3 | 45.9 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 14.9 | 104.6 | GO:1990822 | basic amino acid transmembrane transport(GO:1990822) |
| 14.3 | 57.3 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 13.1 | 52.6 | GO:0002774 | Fc receptor mediated inhibitory signaling pathway(GO:0002774) |
| 12.8 | 63.9 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 12.5 | 37.5 | GO:0038156 | interleukin-3-mediated signaling pathway(GO:0038156) |
| 11.7 | 46.8 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 10.8 | 43.0 | GO:0052148 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 9.6 | 57.4 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 9.4 | 37.4 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 8.4 | 109.5 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 8.2 | 24.6 | GO:2000537 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 7.3 | 7.3 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 7.2 | 14.4 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 7.1 | 35.5 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) |
| 6.9 | 27.7 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 6.4 | 25.8 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 6.2 | 37.0 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 6.0 | 30.0 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 5.9 | 77.3 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 5.9 | 23.5 | GO:0072011 | diapedesis(GO:0050904) glomerular endothelium development(GO:0072011) |
| 5.6 | 118.2 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 5.6 | 16.7 | GO:0002892 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) immune complex clearance(GO:0002434) immune complex clearance by monocytes and macrophages(GO:0002436) type II hypersensitivity(GO:0002445) negative regulation of hypersensitivity(GO:0002884) regulation of type II hypersensitivity(GO:0002892) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) |
| 5.6 | 210.9 | GO:0045730 | respiratory burst(GO:0045730) |
| 5.5 | 38.5 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 5.4 | 26.8 | GO:0071725 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 5.3 | 21.3 | GO:0042097 | interleukin-4 biosynthetic process(GO:0042097) regulation of interleukin-4 biosynthetic process(GO:0045402) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 5.2 | 26.0 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 4.9 | 24.7 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 4.8 | 76.7 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 4.5 | 9.0 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 4.3 | 52.1 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 4.1 | 16.6 | GO:0043335 | protein unfolding(GO:0043335) |
| 3.9 | 23.2 | GO:0010266 | response to vitamin B1(GO:0010266) |
| 3.9 | 34.7 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 3.1 | 9.3 | GO:1903722 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) regulation of centriole elongation(GO:1903722) |
| 3.1 | 15.3 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 3.0 | 18.1 | GO:0015755 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 3.0 | 8.9 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 3.0 | 20.7 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 2.9 | 8.8 | GO:1990654 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 2.9 | 49.7 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 2.9 | 8.7 | GO:0014805 | smooth muscle adaptation(GO:0014805) positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 2.9 | 11.5 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 2.9 | 2.9 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 2.9 | 25.7 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 2.8 | 14.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 2.8 | 8.4 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 2.6 | 10.6 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 2.6 | 7.7 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 2.5 | 15.3 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 2.5 | 233.6 | GO:0032945 | negative regulation of mononuclear cell proliferation(GO:0032945) negative regulation of lymphocyte proliferation(GO:0050672) |
| 2.5 | 39.2 | GO:0071800 | podosome assembly(GO:0071800) |
| 2.3 | 9.0 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) quinolinate biosynthetic process(GO:0019805) |
| 2.3 | 9.0 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 2.2 | 11.2 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 2.2 | 15.5 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 2.2 | 8.7 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 2.0 | 6.1 | GO:0070078 | peptidyl-lysine hydroxylation to 5-hydroxy-L-lysine(GO:0018395) histone arginine demethylation(GO:0070077) histone H3-R2 demethylation(GO:0070078) histone H4-R3 demethylation(GO:0070079) |
| 2.0 | 49.9 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 2.0 | 8.0 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 2.0 | 5.9 | GO:0090234 | regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 1.9 | 23.3 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 1.9 | 18.9 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 1.9 | 37.5 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 1.8 | 5.5 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 1.8 | 7.2 | GO:0019086 | late viral transcription(GO:0019086) |
| 1.8 | 22.8 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 1.7 | 29.9 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 1.6 | 21.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 1.6 | 9.7 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 1.6 | 22.5 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 1.6 | 20.8 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 1.6 | 3.1 | GO:0090135 | actin filament branching(GO:0090135) |
| 1.5 | 4.6 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 1.5 | 3.0 | GO:1990637 | response to prolactin(GO:1990637) |
| 1.5 | 19.4 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 1.4 | 8.4 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 1.4 | 2.8 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 1.4 | 12.5 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 1.4 | 8.4 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 1.4 | 8.2 | GO:0051708 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 1.4 | 8.2 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 1.4 | 5.4 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 1.3 | 15.8 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 1.3 | 5.1 | GO:0071351 | interleukin-18-mediated signaling pathway(GO:0035655) cellular response to interleukin-18(GO:0071351) |
| 1.2 | 10.0 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 1.2 | 6.1 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 1.2 | 7.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 1.2 | 30.2 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 1.1 | 11.5 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 1.1 | 11.1 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 1.1 | 3.3 | GO:0034471 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 1.1 | 4.4 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 1.0 | 3.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 1.0 | 3.1 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) |
| 1.0 | 8.2 | GO:0071569 | protein ufmylation(GO:0071569) |
| 1.0 | 9.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 1.0 | 2.0 | GO:0051462 | cortisol secretion(GO:0043400) regulation of cortisol secretion(GO:0051462) |
| 1.0 | 8.1 | GO:0002353 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 1.0 | 38.3 | GO:0006505 | GPI anchor metabolic process(GO:0006505) GPI anchor biosynthetic process(GO:0006506) |
| 1.0 | 23.7 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 1.0 | 2.9 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.9 | 9.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.9 | 6.5 | GO:0033004 | negative regulation of mast cell activation(GO:0033004) |
| 0.9 | 15.5 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.9 | 18.9 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.9 | 5.3 | GO:0031438 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.9 | 16.7 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.9 | 152.6 | GO:0038096 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.8 | 4.2 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.8 | 9.2 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.8 | 4.0 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.8 | 21.0 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.8 | 4.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) histone H2A phosphorylation(GO:1990164) |
| 0.8 | 4.0 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.8 | 4.7 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.8 | 4.7 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.8 | 23.3 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.8 | 31.4 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.8 | 48.3 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.7 | 6.7 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.7 | 9.6 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.7 | 3.7 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.7 | 7.2 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.7 | 4.9 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.7 | 3.3 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.6 | 4.5 | GO:0051344 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.6 | 4.4 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.6 | 5.6 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.6 | 9.3 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) |
| 0.6 | 2.5 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.6 | 11.3 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.6 | 59.7 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.6 | 4.0 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.6 | 13.5 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.6 | 1.7 | GO:0019605 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 0.6 | 1.7 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.6 | 2.2 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 0.5 | 3.8 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.5 | 7.1 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.5 | 17.0 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.5 | 8.3 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.5 | 57.1 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.5 | 4.1 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.5 | 3.6 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.5 | 1.5 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.5 | 9.1 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.5 | 209.7 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.5 | 6.2 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.5 | 2.8 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.5 | 3.7 | GO:1902661 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) positive regulation of glucose mediated signaling pathway(GO:1902661) |
| 0.5 | 14.5 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
| 0.5 | 3.2 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.5 | 4.1 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.5 | 27.5 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.4 | 7.5 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.4 | 8.2 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.4 | 1.3 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 0.4 | 1.2 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.4 | 72.3 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.4 | 4.3 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.4 | 22.7 | GO:0007077 | mitotic nuclear envelope disassembly(GO:0007077) |
| 0.4 | 7.6 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.4 | 18.0 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.4 | 1.5 | GO:0061042 | adiponectin-activated signaling pathway(GO:0033211) vascular wound healing(GO:0061042) |
| 0.4 | 5.1 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.4 | 7.4 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.3 | 15.6 | GO:0046427 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.3 | 20.7 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.3 | 4.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.3 | 5.4 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.3 | 61.2 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.3 | 3.0 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.3 | 37.4 | GO:0002819 | regulation of adaptive immune response(GO:0002819) |
| 0.3 | 4.5 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.3 | 2.6 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.3 | 1.3 | GO:0036480 | neuron intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0036480) regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903376) |
| 0.3 | 2.2 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.3 | 2.4 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.3 | 0.9 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.3 | 28.7 | GO:0015992 | proton transport(GO:0015992) |
| 0.3 | 3.8 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.3 | 2.3 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.3 | 10.4 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.3 | 1.1 | GO:0001767 | establishment of lymphocyte polarity(GO:0001767) |
| 0.3 | 1.6 | GO:0051958 | methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.3 | 17.1 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.3 | 4.4 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.3 | 5.4 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.2 | 4.9 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.2 | 12.3 | GO:0038034 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.2 | 0.9 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.2 | 3.0 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.2 | 1.4 | GO:0006290 | pyrimidine dimer repair(GO:0006290) activation of JNKK activity(GO:0007256) |
| 0.2 | 2.7 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.2 | 1.3 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) |
| 0.2 | 9.2 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.2 | 2.0 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.2 | 3.3 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.2 | 3.9 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.2 | 6.0 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.2 | 1.5 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.2 | 5.0 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.2 | 9.4 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.2 | 4.7 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.2 | 2.9 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.2 | 5.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.2 | 4.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.2 | 0.9 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.2 | 11.5 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.2 | 11.2 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.2 | 3.1 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.1 | 1.9 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 4.0 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 2.6 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.1 | 2.1 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 4.2 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.1 | 2.7 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 11.8 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.1 | 17.0 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.1 | 1.3 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 3.8 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 68.5 | GO:0043312 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.1 | 1.4 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 0.3 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.1 | 1.2 | GO:0000963 | mitochondrial RNA processing(GO:0000963) |
| 0.1 | 1.1 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.1 | 23.6 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.1 | 0.7 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 14.4 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 0.4 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.1 | 5.8 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.5 | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
| 0.1 | 0.5 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.1 | 1.0 | GO:0015846 | polyamine transport(GO:0015846) |
| 0.1 | 1.7 | GO:0043101 | purine nucleobase metabolic process(GO:0006144) purine-containing compound salvage(GO:0043101) |
| 0.1 | 1.5 | GO:0003299 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.1 | 3.4 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.1 | 0.9 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 1.1 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.1 | 1.3 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 3.4 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 6.2 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.1 | 3.3 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.1 | 2.2 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 0.5 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 1.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 0.5 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 | 0.7 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.1 | 0.3 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.1 | 0.5 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.1 | 6.0 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.6 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 7.5 | GO:0048839 | inner ear development(GO:0048839) |
| 0.1 | 1.8 | GO:0045840 | positive regulation of mitotic nuclear division(GO:0045840) |
| 0.1 | 0.1 | GO:2000157 | negative regulation of protein deubiquitination(GO:0090086) regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.1 | 0.2 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.1 | 3.1 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 5.5 | GO:0044782 | cilium organization(GO:0044782) |
| 0.0 | 1.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 1.5 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 1.2 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.4 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 3.4 | GO:0071774 | response to fibroblast growth factor(GO:0071774) |
| 0.0 | 6.3 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 0.2 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 2.0 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 1.1 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.5 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.0 | 4.7 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.2 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 1.3 | GO:0035113 | embryonic limb morphogenesis(GO:0030326) embryonic appendage morphogenesis(GO:0035113) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 20.2 | 181.8 | GO:0032010 | phagolysosome(GO:0032010) |
| 18.7 | 93.6 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 7.5 | 97.0 | GO:0031209 | SCAR complex(GO:0031209) |
| 7.3 | 188.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 6.9 | 48.4 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 6.6 | 72.4 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 6.2 | 24.8 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 5.4 | 27.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 5.1 | 45.9 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 5.0 | 30.0 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 4.8 | 24.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 4.1 | 24.7 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 3.4 | 17.0 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 3.2 | 38.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 3.1 | 59.4 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 2.6 | 7.7 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 2.6 | 35.8 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 2.5 | 46.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 2.4 | 21.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 2.3 | 47.4 | GO:0097342 | ripoptosome(GO:0097342) |
| 2.2 | 6.5 | GO:0055087 | Ski complex(GO:0055087) |
| 2.1 | 6.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 2.1 | 8.2 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 2.0 | 26.0 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 1.9 | 9.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 1.9 | 23.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 1.8 | 24.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 1.8 | 234.9 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 1.7 | 6.9 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 1.7 | 8.4 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 1.7 | 110.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 1.5 | 106.0 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 1.4 | 11.5 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 1.4 | 5.4 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 1.2 | 3.6 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 1.2 | 16.8 | GO:0033179 | proton-transporting two-sector ATPase complex, proton-transporting domain(GO:0033177) proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 1.2 | 7.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 1.2 | 3.5 | GO:0031251 | PAN complex(GO:0031251) |
| 1.1 | 88.3 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 1.1 | 4.4 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 1.1 | 13.8 | GO:0030897 | HOPS complex(GO:0030897) |
| 1.0 | 79.9 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 1.0 | 4.0 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 1.0 | 4.9 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.9 | 16.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.9 | 6.0 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.8 | 1.7 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.8 | 13.9 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.8 | 99.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.8 | 9.3 | GO:0090543 | Flemming body(GO:0090543) |
| 0.8 | 40.7 | GO:0019814 | immunoglobulin complex(GO:0019814) immunoglobulin complex, circulating(GO:0042571) |
| 0.7 | 27.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.7 | 5.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.7 | 15.5 | GO:0042627 | chylomicron(GO:0042627) |
| 0.7 | 186.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.7 | 31.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.6 | 69.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.6 | 45.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.6 | 8.7 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.6 | 33.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.6 | 2.9 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.6 | 18.5 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.6 | 5.5 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.5 | 4.2 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.5 | 1.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.5 | 5.0 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.5 | 30.0 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.5 | 22.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.5 | 4.5 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.4 | 3.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.4 | 2.0 | GO:0043196 | varicosity(GO:0043196) |
| 0.4 | 27.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.3 | 2.4 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.3 | 1.7 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.3 | 17.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.3 | 10.0 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.3 | 6.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.3 | 46.2 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.3 | 2.3 | GO:0070522 | nucleotide-excision repair factor 1 complex(GO:0000110) ERCC4-ERCC1 complex(GO:0070522) |
| 0.3 | 0.8 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.3 | 2.7 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.3 | 1.0 | GO:0000801 | central element(GO:0000801) |
| 0.3 | 21.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.3 | 8.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.3 | 19.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.3 | 10.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 2.4 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.2 | 4.3 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.2 | 65.0 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.2 | 3.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 11.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.2 | 30.3 | GO:0098802 | plasma membrane receptor complex(GO:0098802) |
| 0.2 | 8.9 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.2 | 67.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.2 | 0.6 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.2 | 41.0 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.2 | 7.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 1.0 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.2 | 3.1 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.2 | 2.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 2.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.2 | 0.7 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.2 | 4.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 40.2 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.2 | 55.2 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.2 | 1.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.2 | 23.1 | GO:0016605 | PML body(GO:0016605) |
| 0.2 | 9.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 10.0 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.2 | 18.2 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.2 | 29.2 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 1.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 12.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 177.5 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 56.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 8.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 7.6 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 3.7 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 1.7 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 11.6 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.1 | 5.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 16.5 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.1 | 3.5 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.7 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 5.1 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 0.5 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 13.1 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.1 | 30.1 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 1.5 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 1.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.2 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.1 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.4 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 2.6 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 4.1 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 3.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 13.8 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 0.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 22.7 | 181.8 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 21.9 | 109.5 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 18.2 | 72.7 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 14.3 | 57.3 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 13.1 | 52.6 | GO:0032396 | inhibitory MHC class I receptor activity(GO:0032396) |
| 12.5 | 37.5 | GO:0004912 | interleukin-3 receptor activity(GO:0004912) |
| 11.9 | 35.8 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 11.9 | 47.7 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 10.9 | 86.9 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 9.8 | 68.4 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 8.7 | 104.6 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 8.2 | 24.7 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 8.1 | 40.5 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 7.2 | 43.0 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 6.9 | 27.7 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 6.8 | 40.7 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 6.7 | 26.8 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 5.7 | 17.0 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 5.5 | 94.1 | GO:0019864 | IgG binding(GO:0019864) |
| 4.9 | 77.6 | GO:0015923 | mannosidase activity(GO:0015923) |
| 4.8 | 14.4 | GO:0048030 | disaccharide binding(GO:0048030) |
| 4.2 | 25.2 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 4.1 | 12.3 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 3.8 | 15.3 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 3.8 | 19.0 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 3.8 | 18.9 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 3.7 | 33.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 3.7 | 51.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 3.3 | 30.0 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 3.3 | 23.2 | GO:0005497 | androgen binding(GO:0005497) |
| 3.3 | 39.6 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 3.0 | 18.1 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 3.0 | 8.9 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 2.9 | 61.9 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 2.9 | 17.5 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 2.9 | 14.5 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 2.9 | 45.9 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 2.8 | 19.9 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 2.6 | 39.6 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 2.6 | 155.9 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 2.5 | 10.0 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 2.5 | 49.9 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 2.5 | 24.6 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 2.5 | 7.4 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 2.4 | 4.9 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 2.3 | 20.8 | GO:0004875 | complement receptor activity(GO:0004875) |
| 2.3 | 20.7 | GO:0043426 | MRF binding(GO:0043426) |
| 2.3 | 9.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 2.2 | 13.2 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 2.2 | 15.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 2.2 | 10.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 2.0 | 6.1 | GO:0033746 | histone demethylase activity (H3-R2 specific)(GO:0033746) histone demethylase activity (H4-R3 specific)(GO:0033749) |
| 2.0 | 12.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 2.0 | 4.0 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 1.9 | 9.5 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 1.8 | 7.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 1.7 | 5.1 | GO:0042008 | interleukin-18 receptor activity(GO:0042008) |
| 1.6 | 13.0 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 1.6 | 8.0 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 1.6 | 48.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 1.6 | 4.7 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 1.5 | 4.6 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 1.5 | 40.2 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 1.5 | 5.9 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 1.4 | 20.7 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 1.4 | 4.1 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 1.4 | 25.8 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 1.3 | 10.7 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 1.3 | 5.1 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 1.3 | 11.5 | GO:0089720 | caspase binding(GO:0089720) |
| 1.3 | 5.0 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 1.2 | 37.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 1.2 | 2.4 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 1.2 | 3.7 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 1.2 | 7.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 1.1 | 15.9 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 1.1 | 15.8 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 1.1 | 5.6 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 1.1 | 11.2 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 1.1 | 6.7 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 1.1 | 3.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 1.1 | 14.8 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 1.0 | 3.1 | GO:0047322 | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity(GO:0047322) [acetyl-CoA carboxylase] kinase activity(GO:0050405) |
| 1.0 | 9.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 1.0 | 11.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 1.0 | 9.0 | GO:0050664 | oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 1.0 | 2.0 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 1.0 | 23.7 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 1.0 | 3.8 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.9 | 11.8 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.9 | 19.6 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.9 | 19.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.9 | 6.2 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.8 | 16.9 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.8 | 13.5 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.8 | 8.7 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.8 | 24.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.8 | 268.0 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.8 | 13.8 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) phosphatidylinositol monophosphate phosphatase activity(GO:0052744) |
| 0.8 | 5.3 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.7 | 22.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.7 | 2.2 | GO:0070039 | rRNA (guanosine-2'-O-)-methyltransferase activity(GO:0070039) |
| 0.7 | 14.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.7 | 11.3 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.7 | 40.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.7 | 9.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.7 | 9.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.7 | 3.5 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.7 | 5.4 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.7 | 25.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.7 | 33.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.6 | 5.7 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.6 | 13.8 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.6 | 44.3 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.6 | 47.0 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.6 | 28.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.6 | 3.5 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.6 | 5.8 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.6 | 1.7 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.6 | 10.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.6 | 9.0 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.6 | 2.8 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.5 | 3.2 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.5 | 20.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.5 | 7.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.5 | 90.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.5 | 1.5 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.5 | 3.9 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.5 | 3.8 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.4 | 2.7 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.4 | 0.9 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.4 | 8.7 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.4 | 1.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.4 | 2.5 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.4 | 12.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.4 | 1.2 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.4 | 4.7 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.4 | 3.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.4 | 8.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.4 | 4.2 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.4 | 5.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.4 | 3.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.4 | 1.1 | GO:0052726 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol tetrakisphosphate kinase activity(GO:0051765) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.4 | 8.2 | GO:0071949 | FAD binding(GO:0071949) |
| 0.3 | 1.7 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.3 | 30.4 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.3 | 4.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.3 | 17.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.3 | 4.4 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.3 | 0.9 | GO:0004040 | amidase activity(GO:0004040) |
| 0.3 | 9.9 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.3 | 45.1 | GO:0003823 | antigen binding(GO:0003823) |
| 0.3 | 8.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.3 | 2.5 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.3 | 2.5 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.3 | 3.6 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.3 | 14.5 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.3 | 6.0 | GO:0046961 | hydrogen-exporting ATPase activity(GO:0036442) ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.3 | 1.6 | GO:0008518 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.2 | 2.5 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.2 | 3.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 20.2 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.2 | 1.5 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.2 | 63.3 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.2 | 28.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 3.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 18.6 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.2 | 3.7 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.2 | 3.8 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.2 | 8.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.2 | 9.0 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.2 | 1.7 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.2 | 38.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.2 | 0.9 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 4.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 29.0 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.1 | 5.5 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 2.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 7.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.5 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.1 | 10.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 3.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 5.4 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.1 | 51.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 3.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 2.0 | GO:0051430 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 1.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 1.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 19.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 8.0 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
| 0.1 | 0.7 | GO:0032407 | MutSalpha complex binding(GO:0032407) |
| 0.1 | 0.9 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 0.4 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.1 | 2.3 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 0.6 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 2.0 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 1.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.3 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 1.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.1 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.6 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 1.2 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.5 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.6 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 1.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.4 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.4 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.5 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 1.2 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 3.9 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.0 | 0.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.4 | GO:0015149 | glucose transmembrane transporter activity(GO:0005355) hexose transmembrane transporter activity(GO:0015149) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 36.7 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 3.7 | 181.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 2.9 | 202.3 | PID EPO PATHWAY | EPO signaling pathway |
| 2.7 | 234.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 2.2 | 34.7 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 2.1 | 47.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 1.9 | 50.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 1.6 | 120.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 1.5 | 19.7 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 1.4 | 28.0 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 1.4 | 67.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 1.3 | 82.7 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 1.2 | 43.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 1.1 | 43.1 | PID BCR 5PATHWAY | BCR signaling pathway |
| 1.1 | 30.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 1.0 | 39.7 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 1.0 | 39.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.7 | 24.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.6 | 16.2 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.6 | 25.5 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.5 | 22.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.5 | 13.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.5 | 21.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.5 | 28.1 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.4 | 27.6 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.4 | 15.3 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.4 | 14.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.4 | 52.0 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.4 | 5.7 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.3 | 7.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 2.7 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.2 | 4.6 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.2 | 2.5 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.2 | 10.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.2 | 4.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.2 | 0.9 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 6.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 1.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 22.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 3.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 3.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 3.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 2.1 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 3.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 4.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 8.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 4.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 32.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 2.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 3.1 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 13.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 2.5 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 9.2 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 0.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 60.9 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 5.6 | 95.9 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 4.5 | 49.6 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 4.3 | 187.7 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 4.3 | 46.8 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 3.4 | 319.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 2.8 | 36.4 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 2.8 | 113.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 2.4 | 17.0 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 2.3 | 109.0 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 2.0 | 24.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 1.9 | 25.0 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 1.6 | 90.6 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 1.3 | 21.1 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 1.3 | 36.7 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 1.2 | 31.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 1.2 | 218.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 1.2 | 15.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 1.1 | 12.4 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.9 | 14.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.9 | 18.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.8 | 4.1 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.8 | 13.9 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.7 | 115.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.7 | 50.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.7 | 11.0 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.6 | 3.1 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.6 | 4.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.6 | 14.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.6 | 21.5 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.6 | 17.0 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.6 | 7.4 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.6 | 35.4 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.5 | 5.9 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.5 | 32.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.5 | 6.8 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.4 | 15.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.4 | 9.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.4 | 5.8 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.3 | 24.4 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.3 | 5.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.3 | 1.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.3 | 6.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.3 | 3.7 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.3 | 5.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 7.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.2 | 6.4 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.2 | 5.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 4.8 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 14.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.2 | 1.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 3.4 | REACTOME CD28 CO STIMULATION | Genes involved in CD28 co-stimulation |
| 0.2 | 18.3 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.2 | 26.5 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.2 | 4.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.2 | 1.5 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.2 | 3.0 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 2.8 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.2 | 3.1 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 3.6 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.2 | 22.3 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.1 | 6.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 3.5 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 4.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 5.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 4.0 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 8.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 5.9 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 6.1 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.1 | 9.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 1.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 5.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 0.9 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 1.4 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 1.5 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |