Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
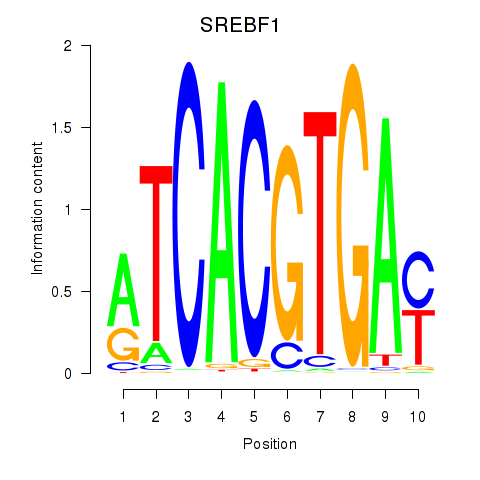
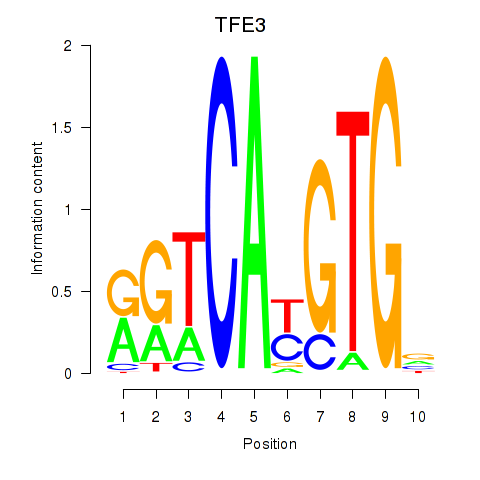
Results for SREBF1_TFE3
Z-value: 2.85
Transcription factors associated with SREBF1_TFE3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SREBF1
|
ENSG00000072310.18 | SREBF1 |
|
TFE3
|
ENSG00000068323.17 | TFE3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TFE3 | hg38_v1_chrX_-_49043345_49043365 | 0.51 | 1.1e-15 | Click! |
| SREBF1 | hg38_v1_chr17_-_17836973_17836993 | 0.01 | 8.5e-01 | Click! |
Activity profile of SREBF1_TFE3 motif
Sorted Z-values of SREBF1_TFE3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SREBF1_TFE3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 19.8 | 59.5 | GO:1902905 | positive regulation of fibril organization(GO:1902905) |
| 14.4 | 43.3 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 12.6 | 75.5 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 11.4 | 34.1 | GO:0061163 | endoplasmic reticulum polarization(GO:0061163) actin filament bundle retrograde transport(GO:0061573) actin filament bundle distribution(GO:0070650) |
| 11.0 | 33.1 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 10.2 | 314.9 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) |
| 9.5 | 28.6 | GO:0007174 | epidermal growth factor catabolic process(GO:0007174) |
| 9.2 | 92.5 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 8.1 | 48.3 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 7.3 | 43.8 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 7.2 | 21.7 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 6.6 | 19.7 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 6.1 | 18.4 | GO:0044830 | modulation by host of viral RNA genome replication(GO:0044830) positive regulation of intracellular transport of viral material(GO:1901254) |
| 6.1 | 30.4 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 5.4 | 21.5 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 5.3 | 15.9 | GO:1904579 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 5.1 | 46.1 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 5.0 | 35.2 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 4.8 | 28.8 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 4.5 | 31.8 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 4.4 | 13.2 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 4.1 | 12.3 | GO:0002339 | B cell selection(GO:0002339) apoptotic process involved in embryonic digit morphogenesis(GO:1902263) regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 4.1 | 16.3 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 4.0 | 44.5 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 4.0 | 16.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 3.9 | 11.8 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 3.6 | 10.9 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 3.5 | 10.4 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 3.4 | 17.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 3.3 | 20.0 | GO:0097338 | response to clozapine(GO:0097338) |
| 3.3 | 9.9 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 3.1 | 9.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 3.0 | 18.1 | GO:1900369 | regulation of RNA interference(GO:1900368) negative regulation of RNA interference(GO:1900369) |
| 3.0 | 12.0 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) |
| 2.9 | 17.6 | GO:1904978 | regulation of endosome organization(GO:1904978) |
| 2.9 | 43.6 | GO:0000338 | protein deneddylation(GO:0000338) |
| 2.9 | 11.6 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 2.9 | 11.5 | GO:0052255 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 2.8 | 19.7 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 2.8 | 30.9 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 2.8 | 33.4 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 2.7 | 8.2 | GO:1901805 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 2.7 | 13.6 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 2.7 | 8.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 2.7 | 32.2 | GO:0070459 | prolactin secretion(GO:0070459) |
| 2.7 | 8.0 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 2.6 | 10.5 | GO:0012502 | induction of programmed cell death(GO:0012502) peptidyl-cysteine S-trans-nitrosylation(GO:0035606) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) |
| 2.5 | 17.8 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 2.5 | 7.4 | GO:0070625 | zymogen granule exocytosis(GO:0070625) |
| 2.4 | 9.8 | GO:0042441 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 2.4 | 19.5 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 2.4 | 14.5 | GO:0006050 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 2.4 | 14.4 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 2.4 | 16.5 | GO:0015811 | L-cystine transport(GO:0015811) |
| 2.3 | 16.4 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 2.3 | 11.7 | GO:0090383 | phagosome acidification(GO:0090383) |
| 2.3 | 15.8 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 2.2 | 6.7 | GO:1902997 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) regulation of neurofibrillary tangle assembly(GO:1902996) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 2.2 | 6.5 | GO:0015808 | L-alanine transport(GO:0015808) |
| 2.1 | 12.7 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 2.1 | 6.3 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) |
| 2.1 | 19.0 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 2.1 | 2.1 | GO:0050787 | detoxification of mercury ion(GO:0050787) |
| 2.0 | 12.1 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 2.0 | 5.9 | GO:0070343 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 2.0 | 15.7 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 1.9 | 40.4 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 1.9 | 84.3 | GO:0006921 | cellular component disassembly involved in execution phase of apoptosis(GO:0006921) |
| 1.9 | 28.3 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 1.9 | 35.7 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 1.9 | 9.3 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 1.9 | 1.9 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 1.8 | 7.3 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 1.8 | 14.4 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 1.7 | 5.0 | GO:1903381 | neuron intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0036483) regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903381) negative regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903382) |
| 1.7 | 16.8 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 1.7 | 3.3 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 1.6 | 13.1 | GO:0070842 | aggresome assembly(GO:0070842) |
| 1.6 | 43.6 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 1.6 | 11.0 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 1.6 | 14.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 1.5 | 4.6 | GO:0097476 | NMDA glutamate receptor clustering(GO:0097114) spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 1.5 | 10.8 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 1.5 | 7.7 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 1.5 | 4.5 | GO:1903565 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 1.5 | 4.5 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 1.5 | 8.8 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 1.5 | 14.6 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 1.5 | 17.5 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 1.4 | 2.9 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 1.4 | 21.6 | GO:0033572 | transferrin transport(GO:0033572) |
| 1.4 | 14.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 1.4 | 4.2 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 1.4 | 15.5 | GO:0006108 | malate metabolic process(GO:0006108) |
| 1.4 | 19.7 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 1.4 | 11.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 1.4 | 5.6 | GO:0070295 | renal water absorption(GO:0070295) |
| 1.4 | 8.2 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 1.4 | 13.6 | GO:0048069 | eye pigmentation(GO:0048069) |
| 1.4 | 19.0 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 1.3 | 13.5 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 1.3 | 18.7 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 1.3 | 9.3 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 1.3 | 4.0 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 1.3 | 5.2 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 1.3 | 9.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 1.3 | 6.5 | GO:0043128 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 1.3 | 6.5 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 1.3 | 7.8 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 1.3 | 5.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 1.3 | 10.1 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 1.2 | 8.7 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 1.2 | 3.7 | GO:0042727 | flavin-containing compound biosynthetic process(GO:0042727) |
| 1.2 | 11.9 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 1.2 | 10.5 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 1.1 | 2.3 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 1.1 | 4.5 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 1.1 | 3.4 | GO:0090382 | phagosome maturation(GO:0090382) |
| 1.1 | 3.3 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 1.1 | 5.3 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 1.1 | 5.3 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 1.1 | 3.2 | GO:0014810 | positive regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014810) |
| 1.0 | 5.2 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 1.0 | 13.2 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 1.0 | 7.0 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 1.0 | 2.9 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 1.0 | 10.6 | GO:0050812 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 1.0 | 3.9 | GO:1903421 | regulation of synaptic vesicle recycling(GO:1903421) |
| 1.0 | 1.9 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 1.0 | 5.7 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.9 | 3.8 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.9 | 5.7 | GO:2000286 | regulation of endosome size(GO:0051036) receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.9 | 2.8 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.9 | 11.2 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.9 | 7.3 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.9 | 23.5 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.9 | 5.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.9 | 8.9 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) positive regulation of ATP biosynthetic process(GO:2001171) |
| 0.9 | 2.6 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.9 | 16.5 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.9 | 6.1 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.9 | 13.8 | GO:1903817 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.8 | 2.5 | GO:0042822 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 biosynthetic process(GO:0042819) pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.8 | 27.0 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.8 | 20.1 | GO:0060742 | epithelial cell differentiation involved in prostate gland development(GO:0060742) |
| 0.8 | 5.8 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.8 | 3.2 | GO:0046618 | drug export(GO:0046618) |
| 0.8 | 1.6 | GO:1903525 | regulation of membrane tubulation(GO:1903525) positive regulation of membrane tubulation(GO:1903527) |
| 0.8 | 9.5 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.8 | 10.3 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.8 | 6.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.8 | 3.9 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.8 | 6.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.8 | 7.7 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.8 | 2.3 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.8 | 0.8 | GO:1902954 | regulation of early endosome to recycling endosome transport(GO:1902954) |
| 0.8 | 22.7 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.8 | 3.8 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.7 | 5.2 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.7 | 3.6 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.7 | 5.0 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.7 | 3.6 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.7 | 25.3 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.7 | 6.3 | GO:0015868 | purine nucleotide transport(GO:0015865) purine ribonucleotide transport(GO:0015868) |
| 0.7 | 3.5 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 0.7 | 2.1 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) |
| 0.7 | 16.6 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.7 | 2.0 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.7 | 5.4 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.7 | 4.7 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.7 | 18.6 | GO:1901985 | positive regulation of protein acetylation(GO:1901985) |
| 0.7 | 7.3 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.7 | 2.0 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.7 | 4.6 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.6 | 10.9 | GO:2000615 | regulation of histone H3-K9 acetylation(GO:2000615) |
| 0.6 | 1.9 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.6 | 5.7 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.6 | 1.3 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.6 | 5.6 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.6 | 1.2 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.6 | 4.3 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.6 | 2.4 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.6 | 77.8 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.6 | 3.6 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.6 | 8.9 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.6 | 5.9 | GO:0006771 | riboflavin metabolic process(GO:0006771) flavin-containing compound metabolic process(GO:0042726) |
| 0.6 | 14.5 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.6 | 9.9 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.6 | 6.9 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.6 | 2.8 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) |
| 0.6 | 1.7 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.6 | 1.7 | GO:2001153 | negative regulation of glycogen (starch) synthase activity(GO:2000466) regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.5 | 4.9 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.5 | 3.2 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.5 | 1.6 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.5 | 6.4 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.5 | 3.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.5 | 7.8 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.5 | 4.1 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.5 | 1.5 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.5 | 1.5 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.5 | 30.2 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.5 | 22.6 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.5 | 53.4 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.5 | 7.8 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.5 | 2.4 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.5 | 20.7 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.5 | 12.9 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.5 | 2.4 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.5 | 4.8 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.5 | 8.1 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.5 | 12.7 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.5 | 48.1 | GO:0015992 | hydrogen transport(GO:0006818) proton transport(GO:0015992) |
| 0.5 | 1.4 | GO:0021779 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.5 | 3.2 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.4 | 18.3 | GO:1904031 | positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.4 | 14.9 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.4 | 2.9 | GO:0044362 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.4 | 123.7 | GO:0006457 | protein folding(GO:0006457) |
| 0.4 | 1.6 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.4 | 1.2 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.4 | 9.5 | GO:0007567 | parturition(GO:0007567) |
| 0.4 | 1.6 | GO:0043485 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.4 | 7.7 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.4 | 3.5 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.4 | 28.5 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.4 | 2.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.4 | 1.4 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.4 | 10.2 | GO:0009268 | response to pH(GO:0009268) |
| 0.3 | 3.5 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.3 | 4.8 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.3 | 1.7 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.3 | 1.7 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.3 | 2.3 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.3 | 2.0 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.3 | 8.9 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 1.6 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.3 | 8.8 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.3 | 0.6 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 0.3 | 1.9 | GO:0032906 | transforming growth factor beta2 production(GO:0032906) regulation of transforming growth factor beta2 production(GO:0032909) |
| 0.3 | 2.8 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.3 | 1.4 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.3 | 1.1 | GO:1990314 | cellular response to insulin-like growth factor stimulus(GO:1990314) |
| 0.3 | 1.4 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.3 | 0.8 | GO:1903012 | positive regulation of bone development(GO:1903012) |
| 0.3 | 5.6 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.3 | 1.6 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.3 | 4.0 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.3 | 2.6 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.3 | 9.0 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.3 | 2.9 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.3 | 8.3 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.3 | 4.9 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.3 | 2.6 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.2 | 5.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 0.7 | GO:0061155 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm development(GO:0048389) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) cloacal septation(GO:0060197) bud dilation involved in lung branching(GO:0060503) regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) pattern specification involved in mesonephros development(GO:0061227) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in kidney development(GO:0072098) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.2 | 5.8 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.2 | 0.2 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.2 | 4.3 | GO:0090190 | positive regulation of mesonephros development(GO:0061213) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.2 | 2.1 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.2 | 14.3 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.2 | 1.3 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.2 | 1.3 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.2 | 0.6 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.2 | 6.4 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.2 | 1.5 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.2 | 5.7 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.2 | 5.7 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.2 | 1.0 | GO:1901091 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.2 | 1.8 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.2 | 2.0 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.2 | 59.2 | GO:0010038 | response to metal ion(GO:0010038) |
| 0.2 | 1.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.2 | 11.6 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.2 | 5.0 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.2 | 1.3 | GO:0015886 | heme transport(GO:0015886) |
| 0.2 | 2.9 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.2 | 1.4 | GO:2000727 | positive regulation of cardiac muscle cell differentiation(GO:2000727) |
| 0.2 | 4.1 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.2 | 6.1 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.2 | 2.6 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.2 | 2.8 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.2 | 4.3 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.2 | 2.4 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.2 | 2.4 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.2 | 0.8 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.2 | 0.8 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.2 | 0.8 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.2 | 0.8 | GO:0042423 | catechol-containing compound biosynthetic process(GO:0009713) catecholamine biosynthetic process(GO:0042423) |
| 0.2 | 1.5 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.2 | 2.1 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.9 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.1 | 2.2 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.1 | 1.9 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.1 | 4.4 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.1 | 0.4 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.1 | 1.5 | GO:0014887 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.1 | 0.4 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.1 | 0.5 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.1 | 0.5 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.1 | 2.3 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.1 | 0.8 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 5.6 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 3.1 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.1 | 0.9 | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.1 | 5.7 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.1 | 0.7 | GO:0071476 | hypotonic salinity response(GO:0042539) cellular hypotonic response(GO:0071476) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 0.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 1.5 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.1 | 2.2 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 0.6 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.1 | 0.6 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.1 | 0.3 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.1 | 9.0 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 4.8 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.1 | 1.6 | GO:1903432 | regulation of TORC1 signaling(GO:1903432) |
| 0.1 | 3.1 | GO:0043039 | tRNA aminoacylation for protein translation(GO:0006418) amino acid activation(GO:0043038) tRNA aminoacylation(GO:0043039) |
| 0.1 | 4.1 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.1 | 1.3 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.1 | 1.4 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.1 | 0.9 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.1 | 1.6 | GO:0006084 | acetyl-CoA metabolic process(GO:0006084) |
| 0.1 | 2.9 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.1 | 0.6 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 1.2 | GO:0051969 | regulation of transmission of nerve impulse(GO:0051969) |
| 0.1 | 1.2 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.1 | 3.2 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 18.5 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.1 | 4.0 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 2.8 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 2.2 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 0.2 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.1 | 2.7 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.1 | 4.1 | GO:0008344 | adult locomotory behavior(GO:0008344) |
| 0.1 | 2.7 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 0.5 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 1.5 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 0.3 | GO:0040034 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.1 | 3.5 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.1 | 9.8 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 2.8 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.1 | 0.2 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.1 | 1.5 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 1.2 | GO:1904376 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.1 | 3.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 1.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 4.1 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.4 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.1 | 0.4 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 0.6 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 0.4 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 1.0 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 7.9 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 2.1 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 3.5 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 2.0 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.7 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.2 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.3 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 2.2 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 1.4 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.0 | 5.2 | GO:0019216 | regulation of lipid metabolic process(GO:0019216) |
| 0.0 | 0.7 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 1.1 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 0.1 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 1.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.3 | GO:0033866 | coenzyme A metabolic process(GO:0015936) coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.0 | 0.6 | GO:0007618 | mating(GO:0007618) |
| 0.0 | 0.2 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.5 | GO:0051930 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 | 1.0 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.0 | 1.1 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 1.3 | GO:0060348 | bone development(GO:0060348) |
| 0.0 | 0.8 | GO:0030593 | neutrophil chemotaxis(GO:0030593) |
| 0.0 | 1.5 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 1.4 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.0 | 0.6 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.1 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.0 | 0.1 | GO:0030638 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 2.2 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 0.4 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.7 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.0 | 1.2 | GO:0050851 | antigen receptor-mediated signaling pathway(GO:0050851) |
| 0.0 | 0.5 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.4 | GO:0010833 | telomere maintenance via telomere lengthening(GO:0010833) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 22.0 | 88.0 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 11.6 | 92.8 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 11.5 | 46.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 10.5 | 167.8 | GO:0033179 | proton-transporting two-sector ATPase complex, proton-transporting domain(GO:0033177) proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 9.5 | 28.6 | GO:0032419 | extrinsic component of lysosome membrane(GO:0032419) |
| 7.5 | 45.2 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 6.8 | 47.9 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 6.8 | 27.3 | GO:0019034 | viral replication complex(GO:0019034) |
| 4.6 | 18.5 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 4.5 | 58.1 | GO:0097413 | Lewy body(GO:0097413) |
| 3.9 | 27.5 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 3.6 | 21.4 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 3.3 | 13.2 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 3.2 | 22.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 2.7 | 35.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 2.6 | 18.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 2.4 | 9.8 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 2.3 | 9.3 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 2.3 | 9.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 2.0 | 7.8 | GO:0070695 | FHF complex(GO:0070695) |
| 1.8 | 125.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 1.7 | 6.9 | GO:0033263 | CORVET complex(GO:0033263) |
| 1.7 | 6.7 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 1.6 | 13.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 1.6 | 19.0 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 1.5 | 32.5 | GO:0031082 | BLOC complex(GO:0031082) |
| 1.5 | 24.2 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 1.4 | 208.5 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 1.4 | 29.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 1.3 | 33.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 1.3 | 5.2 | GO:1990879 | CST complex(GO:1990879) |
| 1.2 | 19.9 | GO:0042583 | chromaffin granule(GO:0042583) |
| 1.2 | 3.6 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 1.2 | 3.5 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 1.1 | 10.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 1.1 | 12.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 1.1 | 8.9 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 1.1 | 8.7 | GO:0030897 | HOPS complex(GO:0030897) |
| 1.1 | 3.2 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 1.0 | 5.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 1.0 | 2.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 1.0 | 29.0 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 1.0 | 7.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 1.0 | 9.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 1.0 | 8.8 | GO:0045180 | basal cortex(GO:0045180) |
| 1.0 | 20.0 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.9 | 47.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.9 | 2.8 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.9 | 3.6 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.9 | 12.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.9 | 17.8 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.9 | 11.5 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.8 | 43.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.8 | 4.1 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.8 | 4.8 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.8 | 2.4 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.8 | 51.5 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.8 | 3.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.8 | 3.9 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.8 | 25.1 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.7 | 18.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.7 | 10.3 | GO:0032059 | bleb(GO:0032059) |
| 0.7 | 14.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.6 | 17.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.6 | 21.3 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.6 | 11.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.6 | 3.0 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.6 | 8.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.6 | 2.2 | GO:0000801 | central element(GO:0000801) |
| 0.5 | 27.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.5 | 1.5 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.5 | 4.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.5 | 4.0 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.5 | 44.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.5 | 12.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.5 | 112.0 | GO:0005770 | late endosome(GO:0005770) |
| 0.5 | 5.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.5 | 2.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.5 | 2.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.5 | 2.3 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.4 | 5.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.4 | 25.6 | GO:0005776 | autophagosome(GO:0005776) |
| 0.4 | 1.7 | GO:0035363 | histone locus body(GO:0035363) |
| 0.4 | 4.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.4 | 3.0 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.4 | 8.0 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.4 | 4.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.4 | 6.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.4 | 9.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.4 | 4.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.4 | 1.4 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.4 | 9.6 | GO:0043218 | compact myelin(GO:0043218) |
| 0.3 | 35.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.3 | 2.4 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.3 | 14.7 | GO:0016234 | inclusion body(GO:0016234) |
| 0.3 | 3.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.3 | 40.3 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.3 | 10.6 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.3 | 15.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.3 | 10.7 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 0.3 | 35.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.3 | 3.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.3 | 4.7 | GO:0016342 | catenin complex(GO:0016342) |
| 0.3 | 8.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.3 | 58.9 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.3 | 11.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.3 | 3.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.3 | 5.7 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.3 | 18.0 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.2 | 1.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.2 | 21.8 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.2 | 4.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.2 | 0.7 | GO:0055087 | Ski complex(GO:0055087) |
| 0.2 | 1.7 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.2 | 1.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.2 | 13.1 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.2 | 5.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.2 | 35.4 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.2 | 0.6 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.2 | 3.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 2.8 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.2 | 2.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.2 | 2.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 12.9 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.2 | 17.1 | GO:0030175 | filopodium(GO:0030175) |
| 0.2 | 4.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 1.9 | GO:0070938 | contractile ring(GO:0070938) |
| 0.2 | 3.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 6.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 19.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.2 | 0.7 | GO:0071546 | pi-body(GO:0071546) |
| 0.2 | 17.8 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 2.1 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 22.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 13.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 1.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 3.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.8 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.1 | 42.9 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 2.0 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.1 | 2.4 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 6.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 30.0 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 2.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 3.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 14.9 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.1 | 42.5 | GO:0045202 | synapse(GO:0045202) |
| 0.1 | 7.5 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 73.6 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.1 | 6.8 | GO:0005775 | vacuolar lumen(GO:0005775) |
| 0.1 | 1.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.7 | GO:1905202 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.1 | 10.7 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 4.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 254.6 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.1 | 0.6 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 0.5 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 5.8 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 0.5 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 2.0 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 1.6 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 1.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 2.6 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 1.5 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 1.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.3 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 29.1 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 20.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.4 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.2 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 20.1 | 60.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 20.0 | 79.8 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) |
| 17.1 | 51.3 | GO:0031696 | alpha-2C adrenergic receptor binding(GO:0031696) |
| 16.8 | 84.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 15.6 | 62.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 12.9 | 103.0 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 8.6 | 154.1 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) |
| 7.9 | 102.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 7.3 | 43.8 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 7.3 | 29.1 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 7.0 | 35.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 5.7 | 17.2 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 5.4 | 21.7 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 5.1 | 30.6 | GO:0017040 | ceramidase activity(GO:0017040) |
| 4.7 | 42.0 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 4.4 | 13.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 3.8 | 15.3 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 3.5 | 73.8 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 3.5 | 10.5 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 3.4 | 10.3 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 3.3 | 39.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 3.2 | 12.7 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 3.2 | 9.5 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 3.1 | 15.5 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 3.1 | 18.4 | GO:0034046 | poly(G) binding(GO:0034046) |
| 3.0 | 15.0 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 3.0 | 21.0 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 3.0 | 11.8 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 2.8 | 8.5 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 2.8 | 25.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 2.8 | 25.0 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 2.7 | 8.2 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 2.7 | 10.6 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 2.6 | 7.7 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 2.5 | 39.2 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 2.4 | 207.1 | GO:0005507 | copper ion binding(GO:0005507) |
| 2.4 | 24.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 2.4 | 7.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 2.4 | 16.5 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 2.3 | 9.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 2.3 | 16.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 2.3 | 18.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 2.3 | 6.8 | GO:0005135 | erythropoietin receptor binding(GO:0005128) interleukin-3 receptor binding(GO:0005135) |
| 2.2 | 6.7 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 2.2 | 15.5 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 2.2 | 11.0 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 2.2 | 10.9 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 2.1 | 8.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 2.1 | 16.6 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 2.1 | 43.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 2.0 | 61.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 1.9 | 92.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 1.9 | 5.7 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 1.8 | 8.9 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 1.8 | 5.3 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 1.7 | 3.3 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 1.7 | 6.7 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 1.6 | 14.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 1.6 | 9.4 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 1.6 | 40.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 1.5 | 10.4 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 1.5 | 28.0 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 1.4 | 32.4 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 1.3 | 10.7 | GO:0045545 | syndecan binding(GO:0045545) |
| 1.3 | 13.2 | GO:0008430 | selenium binding(GO:0008430) |
| 1.3 | 3.9 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 1.2 | 11.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 1.2 | 3.6 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 1.2 | 3.6 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 1.2 | 13.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 1.2 | 3.6 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 1.2 | 16.3 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 1.2 | 4.6 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 1.2 | 28.8 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 1.1 | 30.4 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 1.1 | 8.9 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 1.1 | 17.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 1.1 | 6.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 1.1 | 5.3 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 1.0 | 88.0 | GO:0019003 | GDP binding(GO:0019003) |
| 1.0 | 14.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 1.0 | 12.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 1.0 | 4.1 | GO:0070905 | serine binding(GO:0070905) |
| 1.0 | 4.0 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 1.0 | 7.8 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.9 | 2.8 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.9 | 18.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.9 | 6.2 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.9 | 0.9 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.9 | 7.0 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.9 | 11.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) steroid hormone binding(GO:1990239) |
| 0.9 | 24.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.8 | 8.9 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.8 | 3.2 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.8 | 6.4 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.8 | 11.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.8 | 28.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.8 | 2.4 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.8 | 4.7 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.7 | 7.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.7 | 1.4 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.7 | 2.8 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.7 | 3.4 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.7 | 5.5 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.7 | 3.4 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.7 | 1.3 | GO:0016154 | thymidine phosphorylase activity(GO:0009032) pyrimidine-nucleoside phosphorylase activity(GO:0016154) |
| 0.6 | 7.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.6 | 3.2 | GO:0045174 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.6 | 9.9 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.6 | 10.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.6 | 4.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.6 | 10.4 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.6 | 3.6 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.6 | 6.5 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.6 | 2.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.6 | 32.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.6 | 17.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.6 | 10.9 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.6 | 25.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.6 | 7.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.6 | 1.7 | GO:0070361 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.5 | 13.6 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.5 | 67.7 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.5 | 9.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.5 | 2.1 | GO:0061714 | methotrexate binding(GO:0051870) folic acid receptor activity(GO:0061714) |
| 0.5 | 6.7 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.5 | 2.0 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.5 | 4.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.5 | 5.5 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.5 | 46.3 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.5 | 19.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.5 | 0.9 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.4 | 2.7 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.4 | 10.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.4 | 15.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.4 | 6.6 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.4 | 21.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.4 | 1.6 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.4 | 10.9 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.4 | 2.8 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.4 | 6.0 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.4 | 4.0 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.4 | 1.2 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.4 | 2.8 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.4 | 27.1 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.4 | 10.7 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.4 | 1.5 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.4 | 4.6 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.4 | 13.6 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.4 | 10.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.4 | 2.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.4 | 2.5 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.3 | 1.0 | GO:0031711 | angiotensin type I receptor activity(GO:0001596) bradykinin receptor binding(GO:0031711) |
| 0.3 | 4.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.3 | 1.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.3 | 4.6 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.3 | 2.9 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.3 | 7.8 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.3 | 1.0 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.3 | 15.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.3 | 3.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.3 | 6.9 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.3 | 20.9 | GO:0017022 | myosin binding(GO:0017022) |
| 0.3 | 20.3 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.3 | 1.4 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.3 | 0.9 | GO:0036505 | prosaposin receptor activity(GO:0036505) |
| 0.3 | 9.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.3 | 1.4 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.3 | 5.7 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.3 | 9.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.3 | 1.0 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.3 | 8.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.3 | 0.8 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.3 | 1.3 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.2 | 4.0 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.2 | 16.4 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.2 | 1.0 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.2 | 1.4 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.2 | 1.4 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.2 | 1.7 | GO:0034485 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.2 | 2.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 3.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 12.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 5.8 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 1.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.2 | 18.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.2 | 0.6 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.2 | 4.5 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.2 | 26.5 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.2 | 2.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.2 | 2.5 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 4.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 7.3 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.2 | 1.4 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.2 | 2.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.2 | 1.6 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 1.0 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.2 | 0.6 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.2 | 1.2 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.2 | 1.4 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.2 | 3.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 0.8 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.2 | 2.8 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.2 | 2.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 1.5 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.2 | 3.9 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 2.5 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.2 | 20.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.2 | 0.5 | GO:0016497 | substance K receptor activity(GO:0016497) |
| 0.2 | 3.8 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.2 | 17.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 3.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.2 | 0.8 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.2 | 0.6 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.2 | 3.7 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 6.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 8.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.6 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 2.0 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 1.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 2.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.4 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.1 | 16.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 0.4 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 0.4 | GO:0070119 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) ciliary neurotrophic factor binding(GO:0070119) |
| 0.1 | 2.5 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 2.9 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.1 | 0.5 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.1 | 2.9 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 1.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 1.7 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 11.7 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.1 | 4.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 1.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.1 | 1.1 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 1.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 1.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 1.5 | GO:0015144 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.1 | 2.7 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 2.0 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.7 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 3.3 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 0.8 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.7 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 6.2 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 0.7 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.1 | 0.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 1.3 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.1 | 24.7 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 2.4 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 4.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 1.7 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 1.1 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.1 | 1.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.2 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 1.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.3 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.3 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.1 | 0.2 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.1 | 0.7 | GO:0032407 | MutSalpha complex binding(GO:0032407) |
| 0.1 | 1.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 1.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 3.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.7 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 8.4 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.7 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 1.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 3.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 2.9 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.4 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 0.1 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 1.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 2.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) ether hydrolase activity(GO:0016803) |
| 0.0 | 0.5 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 30.6 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 1.4 | 62.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 1.1 | 29.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 1.1 | 35.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.7 | 33.5 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.7 | 18.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.7 | 12.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.6 | 31.8 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.6 | 66.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.6 | 39.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.5 | 29.1 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.5 | 150.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.5 | 40.7 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.5 | 3.4 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.5 | 10.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.4 | 32.4 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.4 | 5.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.4 | 11.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.4 | 21.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.4 | 7.1 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.3 | 6.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.3 | 10.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.3 | 16.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.3 | 6.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.3 | 1.5 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.3 | 13.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.3 | 52.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.3 | 4.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.3 | 8.5 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.2 | 5.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.2 | 10.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 17.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.2 | 6.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.2 | 9.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 15.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 9.8 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 3.9 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 10.5 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 5.6 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 2.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 4.1 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 2.6 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 7.1 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.1 | 1.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 1.8 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 0.5 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 1.4 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 2.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 0.4 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 1.4 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 2.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.8 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.4 | 312.1 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 4.0 | 60.7 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 3.4 | 176.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 1.7 | 78.5 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 1.5 | 27.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 1.3 | 10.8 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 1.2 | 32.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 1.2 | 22.6 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 1.2 | 35.0 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 1.1 | 19.2 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 1.1 | 74.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.9 | 100.1 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.8 | 14.6 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.8 | 62.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.8 | 10.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.7 | 15.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.7 | 27.6 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.7 | 20.4 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.6 | 6.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.6 | 55.3 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.6 | 10.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.6 | 17.8 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.6 | 6.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.6 | 7.4 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.6 | 6.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.5 | 107.6 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.5 | 3.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.4 | 16.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.4 | 6.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.4 | 9.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.4 | 10.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.4 | 7.6 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.4 | 13.4 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.3 | 10.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.3 | 3.4 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.3 | 6.0 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.3 | 6.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.3 | 4.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.3 | 5.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 22.6 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.3 | 17.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.3 | 2.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.3 | 3.9 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.3 | 10.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 1.9 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.2 | 6.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 2.9 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 29.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 1.8 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.2 | 2.9 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 3.9 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.2 | 5.1 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.2 | 3.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 3.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 2.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 2.6 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.2 | 10.1 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.2 | 7.6 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.2 | 1.0 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.2 | 8.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.2 | 14.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 2.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 3.1 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 4.7 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.2 | 11.4 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.2 | 3.8 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 3.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 17.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.2 | 2.0 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 1.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.2 | 1.7 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.2 | 5.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 27.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 35.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 5.6 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 10.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.4 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.1 | 2.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 8.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 5.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.5 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 3.3 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 1.7 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 1.9 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 2.8 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 1.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 1.6 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 1.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 3.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 3.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.2 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 0.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 2.1 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.5 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 1.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.1 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |