Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
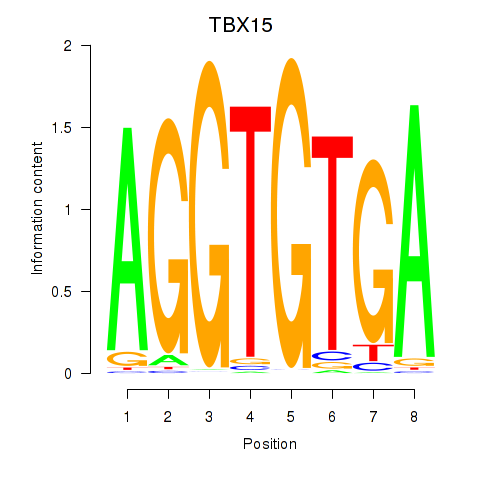
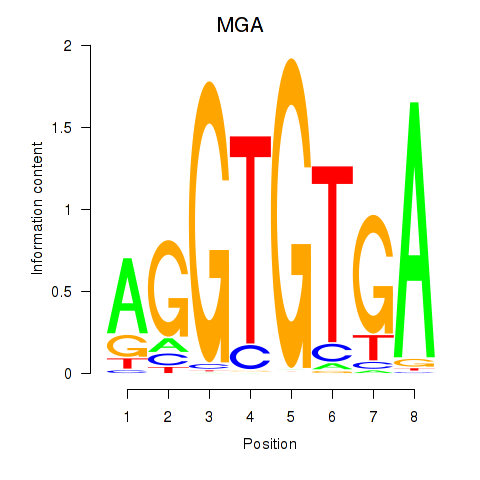
Results for TBX15_MGA
Z-value: 2.03
Transcription factors associated with TBX15_MGA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX15
|
ENSG00000092607.15 | TBX15 |
|
MGA
|
ENSG00000174197.17 | MGA |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MGA | hg38_v1_chr15_+_41660397_41660471 | -0.03 | 6.9e-01 | Click! |
Activity profile of TBX15_MGA motif
Sorted Z-values of TBX15_MGA motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX15_MGA
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 20.0 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 3.0 | 21.0 | GO:0030421 | defecation(GO:0030421) |
| 2.9 | 8.6 | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) |
| 2.6 | 15.3 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 2.5 | 12.7 | GO:1902896 | terminal web assembly(GO:1902896) |
| 2.3 | 11.4 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 2.1 | 12.7 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 2.1 | 8.2 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 2.0 | 12.0 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 1.8 | 5.5 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 1.8 | 10.6 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 1.7 | 8.6 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 1.6 | 4.9 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 1.6 | 6.5 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 1.6 | 4.8 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 1.6 | 4.7 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 1.5 | 9.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 1.4 | 20.2 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 1.4 | 4.3 | GO:0072204 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 1.4 | 15.3 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 1.4 | 31.9 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 1.3 | 14.4 | GO:0035437 | maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 1.2 | 12.0 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 1.2 | 8.3 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 1.2 | 5.9 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 1.1 | 6.8 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 1.1 | 3.2 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 1.0 | 2.1 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 1.0 | 3.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 1.0 | 5.1 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 1.0 | 7.8 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 1.0 | 4.8 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.9 | 3.8 | GO:0035750 | protein localization to myelin sheath abaxonal region(GO:0035750) |
| 0.9 | 2.8 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.9 | 5.5 | GO:0048840 | otolith development(GO:0048840) |
| 0.9 | 4.5 | GO:1901662 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.8 | 5.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.8 | 2.4 | GO:0099542 | trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.8 | 3.0 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.8 | 6.8 | GO:0015865 | purine nucleotide transport(GO:0015865) purine ribonucleotide transport(GO:0015868) |
| 0.7 | 2.2 | GO:1903570 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.7 | 2.2 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.7 | 9.8 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.7 | 2.1 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.7 | 10.3 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.7 | 2.0 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.7 | 2.0 | GO:0060031 | cardiac right atrium morphogenesis(GO:0003213) mediolateral intercalation(GO:0060031) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) regulation of cell proliferation in midbrain(GO:1904933) |
| 0.7 | 1.3 | GO:0060621 | regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 0.7 | 4.6 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.6 | 3.9 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.6 | 2.6 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.6 | 9.0 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.6 | 3.2 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.6 | 1.8 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.6 | 1.7 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.6 | 2.3 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.6 | 1.7 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.5 | 2.2 | GO:2000612 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.5 | 2.7 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.5 | 11.5 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.5 | 16.3 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.5 | 8.1 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.5 | 6.0 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.5 | 4.0 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.5 | 1.9 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.5 | 3.4 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.5 | 6.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.5 | 1.9 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.5 | 1.9 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.5 | 10.7 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.5 | 4.2 | GO:0002069 | columnar/cuboidal epithelial cell maturation(GO:0002069) intestinal epithelial cell maturation(GO:0060574) |
| 0.5 | 1.8 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.5 | 2.3 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.5 | 1.8 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.5 | 4.5 | GO:0021603 | cranial nerve formation(GO:0021603) |
| 0.4 | 1.3 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) response to iron ion starvation(GO:1990641) |
| 0.4 | 1.7 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.4 | 2.5 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.4 | 4.6 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.4 | 2.5 | GO:0070383 | cytidine to uridine editing(GO:0016554) DNA cytosine deamination(GO:0070383) |
| 0.4 | 1.7 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.4 | 1.2 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.4 | 0.4 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.4 | 1.8 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.4 | 1.4 | GO:0006726 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.3 | 5.5 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.3 | 1.3 | GO:0043387 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.3 | 10.0 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.3 | 18.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.3 | 1.6 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.3 | 5.6 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.3 | 2.2 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.3 | 0.9 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.3 | 3.5 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.3 | 12.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.3 | 4.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.3 | 1.6 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.3 | 0.8 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.3 | 1.0 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.3 | 5.9 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.2 | 1.2 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.2 | 1.7 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.2 | 0.7 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.2 | 2.4 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.2 | 4.1 | GO:0048799 | organ maturation(GO:0048799) |
| 0.2 | 0.7 | GO:1904502 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.2 | 2.3 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.2 | 2.5 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.2 | 1.6 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.2 | 3.6 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.2 | 0.9 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.2 | 0.7 | GO:0021834 | embryonic olfactory bulb interneuron precursor migration(GO:0021831) chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) apoptotic process involved in luteolysis(GO:0061364) |
| 0.2 | 1.1 | GO:1903906 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.2 | 0.7 | GO:0046110 | purine nucleobase catabolic process(GO:0006145) xanthine metabolic process(GO:0046110) |
| 0.2 | 1.1 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.2 | 0.7 | GO:0070476 | peptidyl-glutamine methylation(GO:0018364) RNA (guanine-N7)-methylation(GO:0036265) rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.2 | 1.7 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.2 | 2.5 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.2 | 0.8 | GO:0033306 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.2 | 14.5 | GO:0007602 | phototransduction(GO:0007602) |
| 0.2 | 3.8 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.2 | 1.2 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.2 | 1.6 | GO:0009597 | detection of virus(GO:0009597) |
| 0.2 | 5.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 3.0 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.2 | 0.6 | GO:0071934 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.2 | 1.9 | GO:0070170 | regulation of tooth mineralization(GO:0070170) |
| 0.2 | 2.4 | GO:0001767 | establishment of lymphocyte polarity(GO:0001767) |
| 0.2 | 0.7 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.2 | 3.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.2 | 0.9 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.2 | 2.9 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.2 | 0.4 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.2 | 3.5 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.2 | 2.0 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.2 | 3.0 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.2 | 0.5 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.2 | 14.8 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.2 | 3.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 6.7 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.2 | 0.5 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 0.2 | 2.1 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.2 | 1.9 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.2 | 2.0 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 2.5 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 2.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.2 | 1.4 | GO:0002667 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.2 | 2.9 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.1 | 7.1 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.7 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 3.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 5.5 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 2.4 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.1 | 5.1 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.1 | 1.0 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.1 | 4.4 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.1 | 1.4 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 1.6 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 2.9 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 3.2 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 0.4 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.1 | 4.1 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 4.4 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 9.3 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 1.4 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.8 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 1.0 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 2.9 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.1 | 0.3 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.1 | 6.5 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.1 | 1.0 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 6.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.8 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.1 | 0.5 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 | 5.3 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.1 | 3.5 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.1 | 0.9 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 1.7 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 3.2 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.1 | 5.6 | GO:0001658 | branching involved in ureteric bud morphogenesis(GO:0001658) |
| 0.1 | 0.8 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.1 | 2.5 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.1 | 4.7 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.1 | 0.3 | GO:1900226 | positive regulation of oocyte maturation(GO:1900195) negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.1 | 0.4 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 4.7 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.1 | 5.9 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 0.5 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 2.8 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.7 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 1.8 | GO:0071420 | response to histamine(GO:0034776) cellular response to histamine(GO:0071420) |
| 0.1 | 2.0 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 4.9 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.1 | 18.2 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.1 | 2.7 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.1 | 1.4 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.1 | 0.8 | GO:0046149 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 1.4 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.1 | 1.1 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 1.8 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 4.1 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.1 | 4.4 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.3 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 0.3 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 3.9 | GO:1901185 | negative regulation of ERBB signaling pathway(GO:1901185) |
| 0.1 | 1.0 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.9 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 1.2 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.1 | 4.6 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.1 | 0.5 | GO:0046520 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.1 | 0.2 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.1 | 1.1 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.1 | 1.2 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 0.4 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 2.3 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.3 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.0 | 0.3 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 1.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0051039 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.0 | 0.7 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 2.4 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.9 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 0.6 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.3 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 | 0.2 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 2.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 1.0 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 5.5 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.0 | 2.8 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 1.5 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 0.2 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 1.7 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA transport(GO:0051031) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 2.0 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 1.9 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 1.9 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.3 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.9 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.1 | GO:2000504 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) endocardial cushion to mesenchymal transition(GO:0090500) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.8 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.4 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 0.8 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 1.5 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.5 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.4 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 1.3 | GO:0006110 | regulation of glycolytic process(GO:0006110) |
| 0.0 | 2.5 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) 4-hydroxyproline catabolic process(GO:0019470) |
| 0.0 | 2.4 | GO:0030509 | BMP signaling pathway(GO:0030509) |
| 0.0 | 0.2 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 3.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 1.5 | GO:1901016 | regulation of potassium ion transmembrane transporter activity(GO:1901016) |
| 0.0 | 1.2 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.9 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 4.1 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 0.6 | GO:0061053 | somite development(GO:0061053) |
| 0.0 | 3.0 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.0 | 1.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.5 | GO:2000810 | regulation of bicellular tight junction assembly(GO:2000810) |
| 0.0 | 0.2 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.6 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.2 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 1.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.3 | GO:0048536 | spleen development(GO:0048536) |
| 0.0 | 0.3 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.2 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 2.4 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.3 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 25.8 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 2.0 | 11.7 | GO:1990357 | terminal web(GO:1990357) |
| 1.4 | 26.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.9 | 13.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.8 | 4.2 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.8 | 4.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.8 | 12.0 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.7 | 15.0 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.7 | 9.0 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.6 | 5.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.6 | 5.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.6 | 2.5 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.6 | 5.5 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.6 | 3.5 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.6 | 5.7 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.6 | 5.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.5 | 1.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.5 | 1.6 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.5 | 11.6 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.5 | 13.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.4 | 1.7 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.4 | 6.4 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.4 | 4.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.4 | 4.9 | GO:0090543 | Flemming body(GO:0090543) |
| 0.4 | 5.6 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.4 | 5.0 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.4 | 1.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.3 | 2.4 | GO:0043196 | varicosity(GO:0043196) |
| 0.3 | 6.7 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.3 | 7.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.3 | 2.6 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.3 | 2.4 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.3 | 0.8 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.3 | 3.9 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.2 | 1.7 | GO:0044326 | chromaffin granule membrane(GO:0042584) dendritic spine neck(GO:0044326) |
| 0.2 | 22.1 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.2 | 21.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 3.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.2 | 0.9 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.2 | 6.7 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.2 | 5.9 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.2 | 1.5 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.2 | 1.4 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.2 | 2.4 | GO:0071203 | WASH complex(GO:0071203) |
| 0.2 | 4.2 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.2 | 2.5 | GO:0010369 | chromocenter(GO:0010369) |
| 0.2 | 4.0 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.2 | 6.1 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.2 | 1.5 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.2 | 3.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.2 | 36.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.2 | 8.0 | GO:0032420 | stereocilium(GO:0032420) |
| 0.2 | 10.4 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.2 | 0.7 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.2 | 5.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 6.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 0.3 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.2 | 1.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 12.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.2 | 1.3 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.2 | 2.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 2.8 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.1 | 7.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 2.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.8 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 8.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 2.9 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 6.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.7 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.7 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.1 | 4.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.6 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 1.0 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 30.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 6.2 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 1.4 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 4.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 1.4 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 1.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 1.0 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.7 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.5 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 1.0 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 1.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.7 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.1 | 0.9 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.9 | GO:0030677 | ribonuclease P complex(GO:0030677) |
| 0.1 | 2.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.3 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 8.9 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 4.0 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 0.6 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 0.7 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 0.2 | GO:0001940 | female pronucleus(GO:0001939) male pronucleus(GO:0001940) |
| 0.1 | 3.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 2.4 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 2.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.3 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 5.0 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.9 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 6.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.7 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 3.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 2.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 7.0 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 0.8 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.1 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 4.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 2.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.1 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 2.4 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 10.4 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 1.2 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 28.3 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 0.1 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 13.5 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) nuclear outer membrane-endoplasmic reticulum membrane network(GO:0042175) |
| 0.0 | 1.0 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.6 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.9 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 0.3 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.0 | GO:0043204 | perikaryon(GO:0043204) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.8 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 3.3 | 20.0 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 2.5 | 15.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 2.1 | 8.6 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 1.6 | 4.9 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 1.6 | 4.8 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 1.6 | 12.7 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 1.5 | 7.7 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 1.5 | 4.5 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 1.2 | 3.5 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 1.1 | 4.6 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 1.1 | 13.0 | GO:0038132 | neuregulin binding(GO:0038132) |
| 1.0 | 22.9 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 1.0 | 9.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.9 | 6.5 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.9 | 8.9 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.9 | 12.3 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.8 | 12.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.8 | 2.5 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.8 | 4.2 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.8 | 3.0 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.7 | 21.5 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.7 | 2.1 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.7 | 2.8 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.7 | 18.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.7 | 6.0 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.7 | 6.0 | GO:0043426 | MRF binding(GO:0043426) |
| 0.7 | 4.6 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.6 | 1.9 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 0.6 | 12.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.6 | 2.5 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.6 | 2.5 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.6 | 2.4 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.6 | 2.3 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.6 | 2.3 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.5 | 2.5 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.5 | 2.9 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.5 | 6.0 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.5 | 1.8 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.4 | 2.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.4 | 7.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.4 | 2.6 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.4 | 11.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.4 | 2.4 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.4 | 5.9 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.4 | 1.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.4 | 1.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.3 | 1.7 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.3 | 7.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.3 | 4.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.3 | 2.5 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.3 | 32.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.3 | 2.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.3 | 1.2 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.3 | 1.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.3 | 0.8 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.3 | 5.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.3 | 0.8 | GO:0019166 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.3 | 2.2 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.3 | 3.2 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.3 | 1.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.3 | 3.2 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.3 | 1.3 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.3 | 5.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.3 | 2.5 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.2 | 2.2 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.2 | 1.0 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.2 | 2.8 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 0.7 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 1.6 | GO:0098988 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.2 | 1.8 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.2 | 2.8 | GO:0031432 | titin binding(GO:0031432) |
| 0.2 | 2.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.2 | 3.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.2 | 0.6 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.2 | 2.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 5.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 3.9 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.2 | 30.9 | GO:0008201 | heparin binding(GO:0008201) |
| 0.2 | 5.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.2 | 1.6 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.2 | 9.2 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.2 | 3.2 | GO:0016208 | AMP binding(GO:0016208) |
| 0.2 | 1.7 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.2 | 6.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 7.6 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.2 | 4.1 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.2 | 0.8 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 1.7 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.1 | 1.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 1.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 1.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 2.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 1.7 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 1.6 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 6.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.7 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.7 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.1 | 0.3 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.1 | 0.6 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 1.5 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 0.5 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.1 | 1.0 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 2.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 4.3 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 0.5 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 3.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 14.8 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.1 | 6.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.3 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.1 | 0.7 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 3.0 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 2.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 3.4 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 0.3 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.1 | 0.7 | GO:0005347 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) ATP transmembrane transporter activity(GO:0005347) purine nucleotide transmembrane transporter activity(GO:0015216) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 1.0 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 2.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 2.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.9 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 1.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.5 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.4 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 3.4 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 13.7 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.1 | 0.5 | GO:0098960 | postsynaptic neurotransmitter receptor activity(GO:0098960) |
| 0.1 | 1.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 6.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 6.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.4 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 6.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 0.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.3 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 0.3 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.8 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 3.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 1.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.2 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.1 | 3.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 1.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 4.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 1.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 1.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 18.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 1.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 1.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 1.0 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.4 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.1 | 0.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 3.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 5.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.0 | 0.3 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 1.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 1.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 2.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.6 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 3.1 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 1.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.1 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.1 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.0 | 3.2 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 2.2 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 1.2 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 11.8 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 2.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.8 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 3.4 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 4.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 2.7 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 1.2 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 3.1 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 1.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.2 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.4 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 1.3 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.6 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.5 | GO:0030552 | cAMP binding(GO:0030552) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 9.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 1.1 | 28.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.8 | 8.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.7 | 2.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.5 | 13.8 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.5 | 17.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.4 | 27.1 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.4 | 22.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.4 | 25.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.3 | 21.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.3 | 4.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.3 | 13.1 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.3 | 8.9 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.3 | 1.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 2.7 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.2 | 3.8 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.2 | 9.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 6.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 2.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.2 | 5.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.2 | 4.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 1.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 2.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 2.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 5.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 2.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 7.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 24.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 6.1 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 7.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 6.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 13.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 4.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 2.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 3.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 7.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 1.2 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 3.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 3.1 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 4.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 6.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 4.3 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 1.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 0.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 2.5 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 0.8 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 2.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 10.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 2.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.9 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 2.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 2.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.5 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.4 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 15.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 1.4 | 26.0 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 1.0 | 14.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.5 | 12.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.5 | 19.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.5 | 18.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.5 | 17.7 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.5 | 10.7 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.4 | 15.3 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.4 | 17.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.3 | 1.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.3 | 6.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.3 | 4.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.3 | 46.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.3 | 5.5 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.3 | 4.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 6.5 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.2 | 6.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 4.0 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 6.0 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.2 | 3.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.2 | 2.9 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 2.9 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.2 | 6.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.2 | 3.7 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.2 | 3.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 6.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 4.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 3.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 2.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 2.8 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 3.5 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 4.0 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 3.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 4.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 4.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 4.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 3.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 11.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 3.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.8 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 3.6 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 1.7 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 7.7 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.1 | 0.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 4.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 4.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 0.7 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 8.8 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 6.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.7 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 4.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 6.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 1.9 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.1 | 0.7 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 9.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 2.5 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 1.8 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.1 | 2.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 2.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.0 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 2.0 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 1.8 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.9 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.3 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 1.0 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 2.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.6 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.2 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 1.4 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 2.0 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |