Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
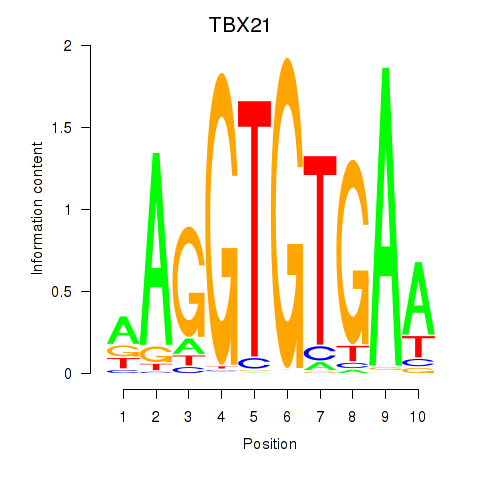
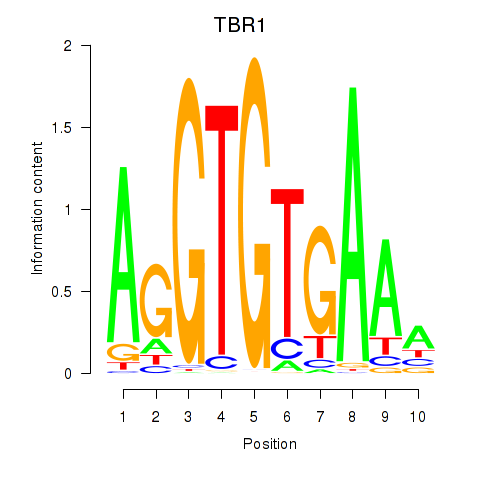
Results for TBX21_TBR1
Z-value: 4.30
Transcription factors associated with TBX21_TBR1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX21
|
ENSG00000073861.3 | TBX21 |
|
TBR1
|
ENSG00000136535.15 | TBR1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX21 | hg38_v1_chr17_+_47733228_47733242 | 0.52 | 2.8e-16 | Click! |
| TBR1 | hg38_v1_chr2_+_161416273_161416325 | 0.25 | 2.2e-04 | Click! |
Activity profile of TBX21_TBR1 motif
Sorted Z-values of TBX21_TBR1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX21_TBR1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 27.1 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 2.9 | 8.7 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 2.7 | 11.0 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 2.6 | 23.3 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 2.3 | 7.0 | GO:0061011 | hepatic duct development(GO:0061011) hepatoblast differentiation(GO:0061017) |
| 1.9 | 11.7 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 1.9 | 7.8 | GO:0090467 | L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 1.9 | 5.8 | GO:0060032 | notochord regression(GO:0060032) |
| 1.8 | 5.4 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 1.7 | 5.2 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 1.7 | 8.6 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 1.5 | 12.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 1.5 | 9.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 1.5 | 4.5 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 1.5 | 4.4 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 1.5 | 4.4 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 1.3 | 5.3 | GO:0002357 | defense response to tumor cell(GO:0002357) |
| 1.2 | 3.7 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 1.2 | 6.0 | GO:0006154 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 1.1 | 3.3 | GO:0060005 | vestibular reflex(GO:0060005) |
| 1.1 | 10.8 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 1.0 | 8.2 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 1.0 | 3.9 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.9 | 1.8 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.9 | 2.7 | GO:0045082 | interleukin-13 biosynthetic process(GO:0042231) positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.9 | 25.8 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.8 | 2.5 | GO:0051039 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.8 | 3.3 | GO:1902617 | response to fluoride(GO:1902617) |
| 0.8 | 2.4 | GO:2000523 | dendritic cell dendrite assembly(GO:0097026) regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.8 | 6.3 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.7 | 3.7 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.7 | 2.8 | GO:0002727 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.7 | 5.3 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.6 | 3.9 | GO:1903282 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) regulation of peroxidase activity(GO:2000468) positive regulation of peroxidase activity(GO:2000470) |
| 0.6 | 5.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.6 | 2.4 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.6 | 2.4 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.6 | 1.7 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.6 | 1.1 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.5 | 1.6 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.5 | 1.6 | GO:0098758 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 0.5 | 8.3 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.5 | 2.7 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.5 | 1.4 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.5 | 1.4 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.4 | 0.9 | GO:0070433 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.4 | 3.5 | GO:0002278 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) regulation of histamine secretion by mast cell(GO:1903593) |
| 0.4 | 0.4 | GO:0045976 | negative regulation of mitotic cell cycle, embryonic(GO:0045976) |
| 0.4 | 8.8 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.4 | 1.2 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 0.4 | 2.7 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.4 | 1.9 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.4 | 1.8 | GO:0044858 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.4 | 1.5 | GO:1900191 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.4 | 3.7 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.4 | 1.8 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.4 | 2.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.4 | 3.2 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.3 | 1.4 | GO:0048627 | myoblast development(GO:0048627) |
| 0.3 | 2.0 | GO:0048541 | positive regulation of gamma-delta T cell differentiation(GO:0045588) mucosal-associated lymphoid tissue development(GO:0048537) Peyer's patch development(GO:0048541) |
| 0.3 | 4.5 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.3 | 2.2 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.3 | 0.9 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.3 | 0.6 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.3 | 0.9 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.3 | 1.2 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.3 | 0.9 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.3 | 1.2 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.3 | 3.0 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.3 | 3.2 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.3 | 8.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 0.9 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.3 | 2.3 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.3 | 0.9 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 0.3 | 4.8 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.3 | 1.7 | GO:0001999 | renal response to blood flow involved in circulatory renin-angiotensin regulation of systemic arterial blood pressure(GO:0001999) renin secretion into blood stream(GO:0002001) |
| 0.3 | 1.7 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.3 | 2.7 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.3 | 3.3 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.3 | 0.8 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.3 | 1.3 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.3 | 3.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.3 | 2.3 | GO:0033089 | negative regulation of T cell mediated cytotoxicity(GO:0001915) positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.2 | 3.0 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.2 | 2.5 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.2 | 1.7 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.2 | 2.0 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.2 | 7.5 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.2 | 2.4 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.2 | 1.2 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.2 | 0.7 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.2 | 3.1 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.2 | 3.3 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 2.6 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.2 | 2.7 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.2 | 1.0 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.2 | 0.4 | GO:0035962 | response to interleukin-13(GO:0035962) |
| 0.2 | 3.5 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 1.0 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.2 | 2.6 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.2 | 0.6 | GO:1904692 | positive regulation of type B pancreatic cell proliferation(GO:1904692) |
| 0.2 | 5.0 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.2 | 1.9 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.2 | 2.5 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.2 | 1.5 | GO:0002353 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.2 | 5.3 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.2 | 2.3 | GO:0015871 | choline transport(GO:0015871) |
| 0.2 | 0.4 | GO:0002331 | pre-B cell differentiation(GO:0002329) pre-B cell allelic exclusion(GO:0002331) |
| 0.2 | 2.0 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.2 | 3.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 0.3 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.2 | 1.6 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.2 | 0.5 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.2 | 0.6 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.2 | 4.0 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.2 | 1.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.2 | 0.5 | GO:1901876 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.2 | 2.3 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 2.5 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 1.0 | GO:0035934 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.1 | 1.3 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.1 | 0.7 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.8 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.1 | 0.8 | GO:0042048 | olfactory behavior(GO:0042048) sensory processing(GO:0050893) |
| 0.1 | 1.9 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 1.3 | GO:0097647 | dimeric G-protein coupled receptor signaling pathway(GO:0038042) calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 1.9 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.1 | 5.0 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 4.6 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.1 | 1.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 4.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 1.6 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 1.6 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.1 | 1.6 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.1 | 3.5 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 1.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 0.5 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.1 | 2.8 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 2.3 | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.1 | 1.6 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 6.9 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 0.5 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.1 | 1.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 1.0 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 0.5 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 2.2 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 0.6 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 0.7 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 1.0 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.1 | 12.7 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.1 | 0.5 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.1 | 0.6 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.1 | 1.8 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.5 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.5 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.1 | 6.8 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.1 | 0.3 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 2.6 | GO:0048536 | spleen development(GO:0048536) |
| 0.1 | 1.9 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 7.3 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.1 | 1.3 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.1 | 1.2 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.1 | 1.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.2 | GO:0018874 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 0.1 | 0.9 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.1 | 2.8 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 1.8 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.0 | 0.4 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 3.3 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 2.2 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.0 | 5.2 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 3.7 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 1.1 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.3 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 5.3 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 0.6 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 1.6 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 1.5 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 1.1 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 3.2 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.0 | 2.4 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 2.2 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 16.7 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 1.2 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 2.0 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.6 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 1.1 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.6 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.6 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 2.0 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.0 | 0.5 | GO:0006349 | regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.0 | 0.6 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 1.0 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 1.1 | GO:0021904 | dorsal/ventral neural tube patterning(GO:0021904) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.4 | GO:0048485 | sympathetic nervous system development(GO:0048485) |
| 0.0 | 0.2 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 1.5 | GO:0071346 | cellular response to interferon-gamma(GO:0071346) |
| 0.0 | 0.9 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 1.0 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 1.0 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.9 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 3.5 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.5 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 1.0 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.0 | 1.1 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.0 | 15.8 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.0 | 0.1 | GO:0072023 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 1.6 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 1.7 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 1.0 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 1.6 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 | 0.5 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.0 | 0.3 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.2 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 0.3 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.6 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 1.3 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.2 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 4.0 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 1.7 | GO:0002262 | myeloid cell homeostasis(GO:0002262) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.8 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 0.3 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.5 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 0.2 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.2 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.3 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.8 | GO:0030574 | collagen catabolic process(GO:0030574) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 23.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 1.5 | 8.8 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 1.1 | 3.3 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.9 | 2.7 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.9 | 3.5 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.6 | 5.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.5 | 5.3 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.5 | 4.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.5 | 1.4 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.4 | 10.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.4 | 11.9 | GO:0032982 | myosin filament(GO:0032982) |
| 0.4 | 1.7 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.4 | 15.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.4 | 2.7 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.4 | 3.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.3 | 4.5 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.3 | 2.4 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.3 | 2.7 | GO:0070187 | telosome(GO:0070187) |
| 0.3 | 3.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.3 | 0.9 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.3 | 0.9 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.3 | 4.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.3 | 2.0 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.3 | 5.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.3 | 1.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.3 | 3.4 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.3 | 1.5 | GO:0005927 | muscle tendon junction(GO:0005927) cuticular plate(GO:0032437) |
| 0.2 | 20.9 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.2 | 3.7 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.2 | 1.3 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.2 | 2.7 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.2 | 0.6 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.2 | 2.6 | GO:0005883 | neurofilament(GO:0005883) |
| 0.2 | 2.9 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 2.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 0.8 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.2 | 5.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.2 | 12.3 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.1 | 15.8 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.1 | 1.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 14.4 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 8.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.8 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 3.4 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 3.9 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.3 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.1 | 0.8 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 1.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.7 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 1.2 | GO:0000124 | SAGA complex(GO:0000124) STAGA complex(GO:0030914) |
| 0.1 | 1.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 1.0 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 2.4 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.1 | 2.6 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 0.6 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 11.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 0.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 2.0 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 4.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 8.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.2 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 5.6 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 8.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 1.5 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 1.6 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 3.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 4.1 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.7 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 1.6 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 2.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.2 | GO:0071664 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 2.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 3.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 4.2 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 6.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.9 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 2.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.6 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.3 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 2.4 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 3.3 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.2 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 1.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 12.2 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.5 | 25.4 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 6.9 | 27.5 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 2.8 | 11.3 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 2.7 | 8.2 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 1.8 | 25.8 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 1.8 | 9.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 1.8 | 5.4 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 1.4 | 8.6 | GO:0003796 | lysozyme activity(GO:0003796) |
| 1.2 | 6.2 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.9 | 6.4 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.7 | 7.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.7 | 3.5 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.7 | 6.0 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.6 | 3.9 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.6 | 2.6 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.6 | 12.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.6 | 3.5 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.6 | 4.5 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.6 | 3.3 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.5 | 11.9 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.5 | 1.6 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.4 | 1.3 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.4 | 26.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.4 | 1.3 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.4 | 4.2 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.4 | 5.8 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.4 | 10.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.4 | 1.1 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.4 | 7.0 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.3 | 2.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.3 | 1.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.3 | 11.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.3 | 1.5 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.3 | 1.2 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.3 | 0.9 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 0.3 | 0.8 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.3 | 7.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 2.7 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.2 | 1.0 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.2 | 5.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 3.9 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.2 | 8.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 2.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 3.5 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.2 | 1.6 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.2 | 4.4 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.2 | 1.3 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.2 | 1.9 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.2 | 2.7 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.2 | 4.6 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.2 | 4.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.2 | 3.7 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.2 | 0.6 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.2 | 3.2 | GO:0017136 | NAD-dependent histone deacetylase activity(GO:0017136) |
| 0.2 | 1.9 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 3.3 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.2 | 0.8 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.2 | 3.9 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.2 | 2.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 2.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.2 | 1.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.2 | 0.6 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.2 | 2.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 3.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 7.5 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 3.8 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.1 | 0.6 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 1.0 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.1 | 5.3 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 10.0 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.1 | 0.4 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.1 | 0.5 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 1.8 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.7 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.1 | 1.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 36.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 1.0 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 0.3 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.1 | 0.5 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.1 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 4.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.4 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 10.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 1.6 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 1.7 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 3.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.5 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.1 | 3.2 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 0.7 | GO:0015216 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) ATP transmembrane transporter activity(GO:0005347) purine nucleotide transmembrane transporter activity(GO:0015216) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 0.9 | GO:0043184 | C-X3-C chemokine binding(GO:0019960) vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.1 | 0.9 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 4.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 1.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.7 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 1.4 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 6.5 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 0.6 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.1 | 1.6 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 11.2 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 1.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 8.4 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 1.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 1.4 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.9 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 2.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 2.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 2.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.4 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 1.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.6 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 1.6 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 2.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 1.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 5.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 2.2 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.1 | 0.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 2.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.6 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 1.3 | GO:0008066 | glutamate receptor activity(GO:0008066) |
| 0.0 | 2.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 15.0 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 3.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 2.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.6 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.3 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 1.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 1.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 3.5 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 4.2 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.5 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 1.7 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 1.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 2.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.4 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 1.0 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.9 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 1.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 1.1 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 4.9 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.5 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.6 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 2.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.3 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 24.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.3 | 13.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.3 | 10.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 12.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.2 | 2.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 9.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.2 | 14.7 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.2 | 17.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.2 | 2.8 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.2 | 2.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 11.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 1.0 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 6.0 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 10.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 8.5 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 3.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 8.0 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 1.6 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 5.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 3.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 2.9 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 2.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 2.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 15.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.9 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 6.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.8 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 2.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 8.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 8.9 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.1 | 2.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 2.2 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 2.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 1.1 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.1 | 1.0 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 2.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.1 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 2.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.8 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 2.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 3.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 3.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 25.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.7 | 11.0 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.6 | 28.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.4 | 7.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.3 | 2.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.3 | 5.3 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.2 | 6.1 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 5.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 20.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.2 | 12.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 13.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 3.7 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.2 | 3.5 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.2 | 6.1 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 3.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 3.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 4.4 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 6.0 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 2.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 2.8 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 8.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 3.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 3.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.0 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 3.8 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 1.5 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 2.3 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 3.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 0.8 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 1.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 1.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 1.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 1.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 1.7 | REACTOME SIGNALLING BY NGF | Genes involved in Signalling by NGF |
| 0.1 | 0.9 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 3.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.7 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 4.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 1.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.6 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 1.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 1.7 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 1.6 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.7 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 1.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.0 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 3.0 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |