Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
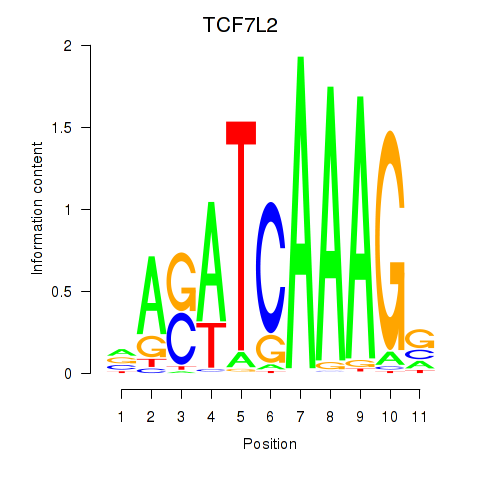
Results for TCF7L2
Z-value: 0.55
Transcription factors associated with TCF7L2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TCF7L2
|
ENSG00000148737.17 | TCF7L2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TCF7L2 | hg38_v1_chr10_+_112950757_112950916 | 0.27 | 4.9e-05 | Click! |
Activity profile of TCF7L2 motif
Sorted Z-values of TCF7L2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TCF7L2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_114257800 | 12.53 |
ENST00000535401.5
|
NNMT
|
nicotinamide N-methyltransferase |
| chr2_-_55917699 | 12.23 |
ENST00000634374.1
|
EFEMP1
|
EGF containing fibulin extracellular matrix protein 1 |
| chr5_-_16936231 | 10.98 |
ENST00000507288.1
ENST00000274203.13 ENST00000513610.6 |
MYO10
|
myosin X |
| chr17_+_1771688 | 8.17 |
ENST00000572048.1
ENST00000573763.1 |
SERPINF1
|
serpin family F member 1 |
| chr2_+_200440649 | 7.51 |
ENST00000366118.2
|
SPATS2L
|
spermatogenesis associated serine rich 2 like |
| chr6_-_11779606 | 6.92 |
ENST00000506810.1
|
ADTRP
|
androgen dependent TFPI regulating protein |
| chr7_+_94394886 | 6.87 |
ENST00000297268.11
ENST00000620463.1 |
COL1A2
|
collagen type I alpha 2 chain |
| chr11_+_69641146 | 6.46 |
ENST00000227507.3
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr16_+_72063226 | 6.21 |
ENST00000540303.7
ENST00000356967.6 ENST00000561690.1 |
HPR
|
haptoglobin-related protein |
| chr5_+_141417659 | 5.92 |
ENST00000398594.4
|
PCDHGB7
|
protocadherin gamma subfamily B, 7 |
| chrX_-_10620419 | 5.91 |
ENST00000380782.6
|
MID1
|
midline 1 |
| chr6_+_31946086 | 5.74 |
ENST00000425368.7
|
CFB
|
complement factor B |
| chrX_-_10620534 | 5.73 |
ENST00000317552.9
|
MID1
|
midline 1 |
| chr10_+_7703340 | 4.94 |
ENST00000429820.5
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr5_+_102865805 | 4.88 |
ENST00000346918.7
|
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr16_+_72054477 | 4.87 |
ENST00000355906.10
ENST00000570083.5 ENST00000228226.12 ENST00000398131.6 ENST00000569639.5 ENST00000564499.5 ENST00000357763.8 ENST00000613898.1 ENST00000562526.5 ENST00000565574.5 ENST00000568417.6 |
HP
|
haptoglobin |
| chr10_+_52314272 | 4.86 |
ENST00000373970.4
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr8_-_118951876 | 4.82 |
ENST00000297350.9
|
TNFRSF11B
|
TNF receptor superfamily member 11b |
| chr15_+_43692886 | 4.71 |
ENST00000434505.5
ENST00000411750.5 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr10_+_7703300 | 4.66 |
ENST00000358415.9
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr13_+_75760659 | 4.41 |
ENST00000526202.5
ENST00000465261.6 |
LMO7
|
LIM domain 7 |
| chr13_+_75760431 | 4.11 |
ENST00000321797.12
|
LMO7
|
LIM domain 7 |
| chr15_+_43593054 | 4.07 |
ENST00000453782.5
ENST00000300283.10 ENST00000437924.5 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr17_-_40100569 | 3.95 |
ENST00000246672.4
|
NR1D1
|
nuclear receptor subfamily 1 group D member 1 |
| chr12_-_12338674 | 3.90 |
ENST00000545735.1
|
MANSC1
|
MANSC domain containing 1 |
| chrX_+_54809060 | 3.89 |
ENST00000396224.1
|
MAGED2
|
MAGE family member D2 |
| chr10_+_99782628 | 3.69 |
ENST00000648689.1
ENST00000647814.1 |
ABCC2
|
ATP binding cassette subfamily C member 2 |
| chr2_+_233712905 | 3.63 |
ENST00000373414.4
|
UGT1A5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr6_-_32178080 | 3.62 |
ENST00000336984.6
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr2_+_69013414 | 3.50 |
ENST00000681816.1
ENST00000482235.2 |
ANTXR1
|
ANTXR cell adhesion molecule 1 |
| chr15_+_64460728 | 3.41 |
ENST00000416172.1
|
ZNF609
|
zinc finger protein 609 |
| chr1_-_93614091 | 3.36 |
ENST00000370247.7
|
BCAR3
|
BCAR3 adaptor protein, NSP family member |
| chr4_+_112860981 | 3.35 |
ENST00000671704.1
|
ANK2
|
ankyrin 2 |
| chr4_+_112860912 | 3.33 |
ENST00000671951.1
|
ANK2
|
ankyrin 2 |
| chr10_+_69801874 | 3.33 |
ENST00000357811.8
|
COL13A1
|
collagen type XIII alpha 1 chain |
| chr1_-_110390989 | 3.30 |
ENST00000369779.9
ENST00000472422.6 |
SLC16A4
|
solute carrier family 16 member 4 |
| chr10_-_73591330 | 3.30 |
ENST00000451492.5
ENST00000681793.1 ENST00000680396.1 ENST00000413442.5 |
USP54
|
ubiquitin specific peptidase 54 |
| chr16_-_73048104 | 3.23 |
ENST00000268489.10
|
ZFHX3
|
zinc finger homeobox 3 |
| chr2_+_69013170 | 3.17 |
ENST00000303714.9
|
ANTXR1
|
ANTXR cell adhesion molecule 1 |
| chr4_+_112861053 | 3.14 |
ENST00000672221.1
|
ANK2
|
ankyrin 2 |
| chr5_-_135954962 | 3.10 |
ENST00000522943.5
ENST00000514447.2 ENST00000274507.6 |
LECT2
|
leukocyte cell derived chemotaxin 2 |
| chr13_-_35855627 | 3.05 |
ENST00000379893.5
|
DCLK1
|
doublecortin like kinase 1 |
| chr1_+_16004228 | 3.03 |
ENST00000329454.2
|
SRARP
|
steroid receptor associated and regulated protein |
| chr5_-_161546671 | 2.99 |
ENST00000517547.5
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr2_+_69013282 | 2.93 |
ENST00000409829.7
|
ANTXR1
|
ANTXR cell adhesion molecule 1 |
| chrX_+_123860262 | 2.91 |
ENST00000355640.3
|
XIAP
|
X-linked inhibitor of apoptosis |
| chrX_+_153494970 | 2.86 |
ENST00000331595.9
ENST00000431891.1 |
BGN
|
biglycan |
| chr10_-_86957582 | 2.85 |
ENST00000372027.10
|
MMRN2
|
multimerin 2 |
| chr2_+_69013379 | 2.81 |
ENST00000409349.7
|
ANTXR1
|
ANTXR cell adhesion molecule 1 |
| chr4_-_138242325 | 2.81 |
ENST00000280612.9
|
SLC7A11
|
solute carrier family 7 member 11 |
| chr17_-_40937445 | 2.77 |
ENST00000436344.7
ENST00000485751.1 |
KRT23
|
keratin 23 |
| chr17_-_40937641 | 2.72 |
ENST00000209718.8
|
KRT23
|
keratin 23 |
| chr13_+_101489940 | 2.66 |
ENST00000376162.7
|
ITGBL1
|
integrin subunit beta like 1 |
| chr10_+_69801892 | 2.63 |
ENST00000398978.8
ENST00000645393.2 ENST00000354547.7 ENST00000674121.1 ENST00000673842.1 ENST00000520267.5 |
COL13A1
|
collagen type XIII alpha 1 chain |
| chr5_+_132257670 | 2.63 |
ENST00000253754.8
ENST00000379018.7 |
PDLIM4
|
PDZ and LIM domain 4 |
| chr6_-_166167832 | 2.63 |
ENST00000366876.7
|
TBXT
|
T-box transcription factor T |
| chr6_-_112254647 | 2.61 |
ENST00000455073.1
ENST00000522006.5 ENST00000519932.5 |
LAMA4
|
laminin subunit alpha 4 |
| chr9_-_23826299 | 2.59 |
ENST00000380117.5
|
ELAVL2
|
ELAV like RNA binding protein 2 |
| chr1_-_110391041 | 2.57 |
ENST00000369781.8
ENST00000437429.6 ENST00000541986.5 |
SLC16A4
|
solute carrier family 16 member 4 |
| chr4_-_108762964 | 2.57 |
ENST00000512646.5
ENST00000411864.6 ENST00000296486.8 ENST00000510706.5 |
ETNPPL
|
ethanolamine-phosphate phospho-lyase |
| chr5_-_161546708 | 2.56 |
ENST00000393959.6
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr13_-_35855758 | 2.54 |
ENST00000615680.4
|
DCLK1
|
doublecortin like kinase 1 |
| chr7_+_27242796 | 2.53 |
ENST00000496902.7
|
EVX1
|
even-skipped homeobox 1 |
| chr5_-_59039454 | 2.52 |
ENST00000358923.10
|
PDE4D
|
phosphodiesterase 4D |
| chr4_-_74099187 | 2.52 |
ENST00000508487.3
|
CXCL2
|
C-X-C motif chemokine ligand 2 |
| chr7_+_138076453 | 2.40 |
ENST00000242375.8
ENST00000411726.6 |
AKR1D1
|
aldo-keto reductase family 1 member D1 |
| chr6_-_112254485 | 2.26 |
ENST00000521398.5
ENST00000424408.6 ENST00000243219.7 |
LAMA4
|
laminin subunit alpha 4 |
| chr8_-_94208548 | 2.20 |
ENST00000027335.8
ENST00000441892.6 ENST00000521491.1 |
CDH17
|
cadherin 17 |
| chr5_-_176388563 | 2.19 |
ENST00000509257.1
ENST00000616685.1 ENST00000614830.5 |
NOP16
|
NOP16 nucleolar protein |
| chr12_+_54033026 | 2.15 |
ENST00000312492.3
|
HOXC5
|
homeobox C5 |
| chr3_+_29281552 | 2.14 |
ENST00000452462.5
ENST00000456853.1 |
RBMS3
|
RNA binding motif single stranded interacting protein 3 |
| chr18_+_48539017 | 2.13 |
ENST00000256413.8
|
CTIF
|
cap binding complex dependent translation initiation factor |
| chr19_-_40285277 | 2.13 |
ENST00000579047.5
ENST00000392038.7 |
AKT2
|
AKT serine/threonine kinase 2 |
| chr2_-_88128049 | 2.13 |
ENST00000393750.3
ENST00000295834.8 |
FABP1
|
fatty acid binding protein 1 |
| chr17_-_69141878 | 2.11 |
ENST00000590645.1
ENST00000284425.7 |
ABCA6
|
ATP binding cassette subfamily A member 6 |
| chr18_+_48539112 | 2.02 |
ENST00000382998.8
|
CTIF
|
cap binding complex dependent translation initiation factor |
| chr8_-_142879820 | 2.02 |
ENST00000377675.3
ENST00000517471.5 ENST00000292427.10 |
CYP11B1
|
cytochrome P450 family 11 subfamily B member 1 |
| chr17_+_44846318 | 2.01 |
ENST00000591513.5
|
HIGD1B
|
HIG1 hypoxia inducible domain family member 1B |
| chr7_+_100015572 | 1.99 |
ENST00000535170.5
|
ZKSCAN1
|
zinc finger with KRAB and SCAN domains 1 |
| chr2_+_11539833 | 1.99 |
ENST00000263834.9
|
GREB1
|
growth regulating estrogen receptor binding 1 |
| chr5_+_161850597 | 1.97 |
ENST00000634335.1
ENST00000635880.1 |
GABRA1
|
gamma-aminobutyric acid type A receptor subunit alpha1 |
| chrX_-_43973382 | 1.96 |
ENST00000642620.1
ENST00000647044.1 |
NDP
|
norrin cystine knot growth factor NDP |
| chr2_+_69013337 | 1.95 |
ENST00000463335.2
|
ANTXR1
|
ANTXR cell adhesion molecule 1 |
| chr9_-_74887720 | 1.91 |
ENST00000449912.6
|
TRPM6
|
transient receptor potential cation channel subfamily M member 6 |
| chr2_+_112911159 | 1.90 |
ENST00000263326.8
|
IL37
|
interleukin 37 |
| chr3_-_134374439 | 1.88 |
ENST00000513145.1
ENST00000249883.10 ENST00000422605.6 |
AMOTL2
|
angiomotin like 2 |
| chr7_+_100015588 | 1.88 |
ENST00000324306.11
ENST00000426572.5 |
ZKSCAN1
|
zinc finger with KRAB and SCAN domains 1 |
| chr5_+_55160161 | 1.88 |
ENST00000296734.6
ENST00000515370.1 ENST00000503787.6 |
GPX8
|
glutathione peroxidase 8 (putative) |
| chr5_-_161546970 | 1.75 |
ENST00000675303.1
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr10_+_113553039 | 1.73 |
ENST00000351270.4
|
HABP2
|
hyaluronan binding protein 2 |
| chr14_+_20768393 | 1.69 |
ENST00000326783.4
|
EDDM3B
|
epididymal protein 3B |
| chrX_+_47078434 | 1.68 |
ENST00000397180.6
|
RGN
|
regucalcin |
| chr6_-_11778781 | 1.66 |
ENST00000414691.8
ENST00000229583.9 |
ADTRP
|
androgen dependent TFPI regulating protein |
| chr9_-_74887914 | 1.62 |
ENST00000360774.6
|
TRPM6
|
transient receptor potential cation channel subfamily M member 6 |
| chr14_-_52791597 | 1.62 |
ENST00000216410.8
ENST00000557604.1 |
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr14_-_52791462 | 1.60 |
ENST00000650397.1
ENST00000554230.5 |
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr3_+_156120572 | 1.59 |
ENST00000389636.9
ENST00000490337.6 |
KCNAB1
|
potassium voltage-gated channel subfamily A member regulatory beta subunit 1 |
| chr12_+_1629197 | 1.57 |
ENST00000397196.7
|
WNT5B
|
Wnt family member 5B |
| chr10_+_47300174 | 1.56 |
ENST00000580279.2
|
GDF10
|
growth differentiation factor 10 |
| chr7_-_75823355 | 1.56 |
ENST00000416943.1
|
CCL24
|
C-C motif chemokine ligand 24 |
| chr2_+_172821575 | 1.55 |
ENST00000397087.7
|
RAPGEF4
|
Rap guanine nucleotide exchange factor 4 |
| chr2_+_172860038 | 1.55 |
ENST00000538974.5
ENST00000540783.5 |
RAPGEF4
|
Rap guanine nucleotide exchange factor 4 |
| chr7_-_27174253 | 1.54 |
ENST00000613671.1
|
HOXA10
|
homeobox A10 |
| chr14_+_74348440 | 1.54 |
ENST00000256362.5
|
VRTN
|
vertebrae development associated |
| chr1_-_11848345 | 1.53 |
ENST00000376476.1
|
NPPA
|
natriuretic peptide A |
| chr19_-_40285395 | 1.53 |
ENST00000424901.5
ENST00000578123.5 |
AKT2
|
AKT serine/threonine kinase 2 |
| chr5_-_24644968 | 1.53 |
ENST00000264463.8
|
CDH10
|
cadherin 10 |
| chr19_-_18606779 | 1.49 |
ENST00000684169.1
ENST00000392386.8 |
CRLF1
|
cytokine receptor like factor 1 |
| chr17_-_47957824 | 1.48 |
ENST00000300557.3
|
PRR15L
|
proline rich 15 like |
| chrX_+_47078380 | 1.48 |
ENST00000352078.8
|
RGN
|
regucalcin |
| chr1_+_99646025 | 1.46 |
ENST00000263174.9
ENST00000605497.5 ENST00000615664.1 |
PALMD
|
palmdelphin |
| chr10_-_102432565 | 1.45 |
ENST00000369937.5
|
CUEDC2
|
CUE domain containing 2 |
| chr11_-_35360050 | 1.44 |
ENST00000644868.1
ENST00000643454.1 ENST00000646080.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr1_+_16367088 | 1.44 |
ENST00000471507.5
ENST00000401089.3 ENST00000401088.9 ENST00000492354.1 |
SZRD1
|
SUZ RNA binding domain containing 1 |
| chr20_+_59300547 | 1.42 |
ENST00000644821.1
|
EDN3
|
endothelin 3 |
| chr11_-_62707413 | 1.42 |
ENST00000360796.10
ENST00000449636.6 |
BSCL2
|
BSCL2 lipid droplet biogenesis associated, seipin |
| chr7_-_151814668 | 1.42 |
ENST00000651764.1
|
PRKAG2
|
protein kinase AMP-activated non-catalytic subunit gamma 2 |
| chr11_-_62707581 | 1.41 |
ENST00000684475.1
ENST00000683296.1 ENST00000684067.1 ENST00000682223.1 |
BSCL2
|
BSCL2 lipid droplet biogenesis associated, seipin |
| chrX_+_47078330 | 1.40 |
ENST00000457380.5
|
RGN
|
regucalcin |
| chr15_-_55588337 | 1.38 |
ENST00000563719.4
|
PYGO1
|
pygopus family PHD finger 1 |
| chr8_-_132111159 | 1.37 |
ENST00000673615.1
ENST00000434736.6 |
HHLA1
|
HERV-H LTR-associating 1 |
| chr7_-_151814636 | 1.36 |
ENST00000652047.1
ENST00000650858.1 |
PRKAG2
|
protein kinase AMP-activated non-catalytic subunit gamma 2 |
| chr1_-_109393197 | 1.36 |
ENST00000538502.5
ENST00000482236.5 |
SORT1
|
sortilin 1 |
| chr4_+_95051671 | 1.32 |
ENST00000440890.7
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chrX_+_15507302 | 1.29 |
ENST00000342014.6
|
BMX
|
BMX non-receptor tyrosine kinase |
| chr1_-_11847772 | 1.27 |
ENST00000376480.7
ENST00000610706.1 |
NPPA
|
natriuretic peptide A |
| chr5_+_67004618 | 1.25 |
ENST00000261569.11
ENST00000436277.5 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr15_-_89221558 | 1.24 |
ENST00000268125.10
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr6_-_112254555 | 1.23 |
ENST00000230538.12
ENST00000389463.9 ENST00000368638.5 ENST00000431543.6 ENST00000453937.2 |
LAMA4
|
laminin subunit alpha 4 |
| chr16_+_19211157 | 1.23 |
ENST00000568433.1
|
SYT17
|
synaptotagmin 17 |
| chr6_-_28252246 | 1.22 |
ENST00000377294.3
|
ZKSCAN4
|
zinc finger with KRAB and SCAN domains 4 |
| chr7_+_111091119 | 1.20 |
ENST00000308478.10
|
LRRN3
|
leucine rich repeat neuronal 3 |
| chr5_-_149551381 | 1.15 |
ENST00000670598.1
ENST00000657001.1 ENST00000515768.6 ENST00000261798.10 ENST00000377843.8 |
CSNK1A1
|
casein kinase 1 alpha 1 |
| chr18_-_26865732 | 1.15 |
ENST00000672188.1
|
AQP4
|
aquaporin 4 |
| chr2_-_201071579 | 1.14 |
ENST00000453765.5
ENST00000452799.5 ENST00000446678.5 ENST00000418596.7 ENST00000681958.1 |
FAM126B
|
family with sequence similarity 126 member B |
| chr1_-_53838276 | 1.08 |
ENST00000371429.4
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chr18_-_26865689 | 1.01 |
ENST00000675739.1
ENST00000383168.9 ENST00000672981.2 ENST00000578776.1 |
AQP4
|
aquaporin 4 |
| chr7_-_80512041 | 1.00 |
ENST00000398291.4
|
GNAT3
|
G protein subunit alpha transducin 3 |
| chr1_+_61952283 | 1.00 |
ENST00000307297.8
|
PATJ
|
PATJ crumbs cell polarity complex component |
| chr7_-_27174274 | 1.00 |
ENST00000283921.5
|
HOXA10
|
homeobox A10 |
| chr14_-_64942783 | 0.99 |
ENST00000612794.1
|
GPX2
|
glutathione peroxidase 2 |
| chr20_+_59300703 | 0.98 |
ENST00000395654.3
|
EDN3
|
endothelin 3 |
| chr10_+_52128343 | 0.95 |
ENST00000672084.1
|
PRKG1
|
protein kinase cGMP-dependent 1 |
| chr3_-_57165332 | 0.95 |
ENST00000296318.12
|
IL17RD
|
interleukin 17 receptor D |
| chr5_-_16742221 | 0.93 |
ENST00000505695.5
|
MYO10
|
myosin X |
| chr20_-_17531366 | 0.93 |
ENST00000377873.8
|
BFSP1
|
beaded filament structural protein 1 |
| chr1_-_162023826 | 0.90 |
ENST00000294794.8
|
OLFML2B
|
olfactomedin like 2B |
| chr8_+_32721823 | 0.87 |
ENST00000539990.3
ENST00000519240.5 |
NRG1
|
neuregulin 1 |
| chr14_+_32329341 | 0.87 |
ENST00000557354.5
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A-kinase anchoring protein 6 |
| chr6_-_25930611 | 0.86 |
ENST00000360488.7
|
SLC17A2
|
solute carrier family 17 member 2 |
| chr5_+_175861628 | 0.86 |
ENST00000509837.5
|
CPLX2
|
complexin 2 |
| chr20_-_57265738 | 0.85 |
ENST00000433911.1
|
BMP7
|
bone morphogenetic protein 7 |
| chr16_+_50548387 | 0.85 |
ENST00000268459.6
|
NKD1
|
NKD inhibitor of WNT signaling pathway 1 |
| chr6_-_25930678 | 0.85 |
ENST00000377850.8
|
SLC17A2
|
solute carrier family 17 member 2 |
| chr7_+_111091006 | 0.84 |
ENST00000451085.5
ENST00000422987.3 ENST00000421101.1 |
LRRN3
|
leucine rich repeat neuronal 3 |
| chr20_+_59300402 | 0.84 |
ENST00000311585.11
ENST00000371028.6 |
EDN3
|
endothelin 3 |
| chr9_-_127950716 | 0.83 |
ENST00000373084.8
|
FAM102A
|
family with sequence similarity 102 member A |
| chr12_+_70366277 | 0.82 |
ENST00000258111.5
|
KCNMB4
|
potassium calcium-activated channel subfamily M regulatory beta subunit 4 |
| chr2_+_48568981 | 0.81 |
ENST00000394754.5
|
STON1-GTF2A1L
|
STON1-GTF2A1L readthrough |
| chr5_-_154478218 | 0.80 |
ENST00000231121.3
|
HAND1
|
heart and neural crest derivatives expressed 1 |
| chr20_+_59300589 | 0.78 |
ENST00000337938.7
ENST00000371025.7 |
EDN3
|
endothelin 3 |
| chr4_-_122621011 | 0.77 |
ENST00000611104.2
ENST00000648588.1 |
IL21
|
interleukin 21 |
| chr17_+_81712236 | 0.76 |
ENST00000545862.5
ENST00000350690.10 ENST00000331531.9 |
SLC25A10
|
solute carrier family 25 member 10 |
| chr12_-_52385649 | 0.73 |
ENST00000257951.3
|
KRT84
|
keratin 84 |
| chr12_-_48570046 | 0.73 |
ENST00000301046.6
ENST00000549817.1 |
LALBA
|
lactalbumin alpha |
| chr14_+_32329256 | 0.72 |
ENST00000280979.9
|
AKAP6
|
A-kinase anchoring protein 6 |
| chr19_-_51082883 | 0.69 |
ENST00000650543.2
|
KLK14
|
kallikrein related peptidase 14 |
| chr10_-_102419934 | 0.61 |
ENST00000406432.5
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr12_+_8914525 | 0.55 |
ENST00000543824.5
|
PHC1
|
polyhomeotic homolog 1 |
| chr3_+_137764296 | 0.55 |
ENST00000306087.3
|
SOX14
|
SRY-box transcription factor 14 |
| chr17_-_41350824 | 0.55 |
ENST00000007735.4
|
KRT33A
|
keratin 33A |
| chr7_+_27242700 | 0.51 |
ENST00000222761.7
|
EVX1
|
even-skipped homeobox 1 |
| chr13_-_27969295 | 0.49 |
ENST00000381020.8
|
CDX2
|
caudal type homeobox 2 |
| chr3_-_177196451 | 0.47 |
ENST00000430069.5
ENST00000630796.2 ENST00000428970.5 |
TBL1XR1
|
TBL1X receptor 1 |
| chr15_-_93089192 | 0.44 |
ENST00000329082.11
|
RGMA
|
repulsive guidance molecule BMP co-receptor a |
| chr1_-_6419903 | 0.42 |
ENST00000377836.8
ENST00000487437.5 ENST00000489730.1 ENST00000377834.8 |
HES2
|
hes family bHLH transcription factor 2 |
| chr10_-_60141004 | 0.41 |
ENST00000355288.6
|
ANK3
|
ankyrin 3 |
| chr5_-_176388629 | 0.39 |
ENST00000619979.4
ENST00000621444.4 |
NOP16
|
NOP16 nucleolar protein |
| chr6_+_63571702 | 0.38 |
ENST00000672924.1
|
PTP4A1
|
protein tyrosine phosphatase 4A1 |
| chr1_-_19923617 | 0.35 |
ENST00000375116.3
|
PLA2G2E
|
phospholipase A2 group IIE |
| chr12_-_23584600 | 0.34 |
ENST00000396007.6
|
SOX5
|
SRY-box transcription factor 5 |
| chr7_-_106285898 | 0.33 |
ENST00000424768.2
ENST00000681255.1 |
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chrX_-_32155462 | 0.30 |
ENST00000359836.5
ENST00000378707.7 ENST00000541735.5 ENST00000684130.1 ENST00000682238.1 ENST00000620040.5 ENST00000474231.5 |
DMD
|
dystrophin |
| chrX_-_47145035 | 0.28 |
ENST00000276062.8
|
NDUFB11
|
NADH:ubiquinone oxidoreductase subunit B11 |
| chr1_+_147541491 | 0.26 |
ENST00000683836.1
ENST00000234739.8 |
BCL9
|
BCL9 transcription coactivator |
| chr7_-_139777986 | 0.26 |
ENST00000406875.8
|
HIPK2
|
homeodomain interacting protein kinase 2 |
| chr10_-_102419693 | 0.26 |
ENST00000611678.4
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr3_+_141368497 | 0.23 |
ENST00000321464.7
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr18_+_10454584 | 0.22 |
ENST00000355285.10
|
APCDD1
|
APC down-regulated 1 |
| chr9_+_133534697 | 0.22 |
ENST00000651351.2
|
ADAMTSL2
|
ADAMTS like 2 |
| chr18_+_32190015 | 0.21 |
ENST00000581447.1
|
MEP1B
|
meprin A subunit beta |
| chr1_-_160579439 | 0.21 |
ENST00000368054.8
ENST00000368048.7 ENST00000311224.8 ENST00000368051.3 ENST00000534968.5 |
CD84
|
CD84 molecule |
| chr10_+_103245887 | 0.21 |
ENST00000441178.2
|
RPEL1
|
ribulose-5-phosphate-3-epimerase like 1 |
| chr6_+_151809105 | 0.17 |
ENST00000427531.6
|
ESR1
|
estrogen receptor 1 |
| chr2_+_203867764 | 0.16 |
ENST00000648405.2
|
CTLA4
|
cytotoxic T-lymphocyte associated protein 4 |
| chr5_+_171419635 | 0.15 |
ENST00000274625.6
|
FGF18
|
fibroblast growth factor 18 |
| chr8_-_6926066 | 0.12 |
ENST00000297436.3
|
DEFA6
|
defensin alpha 6 |
| chr5_-_149551168 | 0.09 |
ENST00000515748.2
ENST00000606719.6 |
CSNK1A1
|
casein kinase 1 alpha 1 |
| chr2_+_227813834 | 0.08 |
ENST00000358813.5
ENST00000409189.7 |
CCL20
|
C-C motif chemokine ligand 20 |
| chr2_-_232776555 | 0.08 |
ENST00000438786.1
ENST00000233826.4 ENST00000409779.1 |
KCNJ13
|
potassium inwardly rectifying channel subfamily J member 13 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 14.4 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 1.7 | 12.2 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 1.6 | 4.9 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 1.6 | 4.9 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) negative regulation of cell fate specification(GO:0009996) |
| 1.3 | 4.0 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 1.2 | 3.7 | GO:0016999 | antibiotic metabolic process(GO:0016999) detoxification of mercury ion(GO:0050787) |
| 1.2 | 4.9 | GO:0001519 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 1.2 | 3.7 | GO:0097473 | response to high light intensity(GO:0009644) cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) retinal cell apoptotic process(GO:1990009) |
| 1.2 | 8.2 | GO:0071279 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) cellular response to cobalt ion(GO:0071279) |
| 1.1 | 4.6 | GO:1903627 | regulation of calcium-dependent ATPase activity(GO:1903610) negative regulation of calcium-dependent ATPase activity(GO:1903611) regulation of dUTP diphosphatase activity(GO:1903627) positive regulation of dUTP diphosphatase activity(GO:1903629) negative regulation of aminoacyl-tRNA ligase activity(GO:1903631) regulation of leucine-tRNA ligase activity(GO:1903633) negative regulation of leucine-tRNA ligase activity(GO:1903634) |
| 1.0 | 3.0 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 1.0 | 9.8 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.8 | 11.6 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.8 | 2.4 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.8 | 12.9 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.7 | 2.2 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.7 | 4.4 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.7 | 2.9 | GO:1902530 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.7 | 4.9 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.6 | 3.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.6 | 3.2 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.6 | 6.5 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.6 | 4.0 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.6 | 3.9 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.5 | 6.9 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.4 | 2.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.4 | 9.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.3 | 4.7 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.3 | 3.6 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.3 | 1.5 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.3 | 2.9 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.3 | 0.8 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.3 | 2.5 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.3 | 5.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.3 | 0.8 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.3 | 2.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.3 | 1.6 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.3 | 3.6 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.3 | 1.0 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.2 | 0.7 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.2 | 0.9 | GO:0072134 | hyaluranon cable assembly(GO:0036118) nephrogenic mesenchyme morphogenesis(GO:0072134) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) allantois development(GO:1905069) |
| 0.2 | 2.0 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.2 | 0.8 | GO:0019418 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.2 | 3.4 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.2 | 1.3 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.2 | 1.5 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.2 | 2.5 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.2 | 2.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 11.9 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.2 | 4.2 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 2.8 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 1.0 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.1 | 1.4 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 2.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 2.8 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 2.0 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.1 | 7.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 3.6 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 5.6 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 1.3 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 6.1 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 1.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 0.5 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 1.5 | GO:0032196 | transposition(GO:0032196) |
| 0.1 | 2.5 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 1.7 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 2.2 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 0.8 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.1 | 0.9 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 0.8 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 0.2 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.1 | 1.4 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.1 | 7.0 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 1.4 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 0.3 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.1 | 0.5 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.9 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 1.1 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 6.1 | GO:0045995 | regulation of embryonic development(GO:0045995) |
| 0.0 | 1.9 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 6.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.0 | 1.2 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 3.1 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.8 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 4.1 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.2 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 1.6 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.2 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.0 | 0.4 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.9 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 2.6 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.1 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.0 | 2.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.4 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.6 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.0 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 1.2 | 4.9 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.8 | 26.6 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.7 | 8.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.5 | 6.9 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.5 | 1.5 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.4 | 2.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.4 | 6.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.4 | 3.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 1.6 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 9.0 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 1.6 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.2 | 10.2 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 1.1 | GO:0005816 | spindle pole body(GO:0005816) |
| 0.1 | 0.9 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.1 | 2.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 6.1 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 6.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 6.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 1.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 13.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 25.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 6.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 2.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 1.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 2.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 5.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 2.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 4.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 4.0 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 5.6 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 2.9 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 3.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 5.7 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 2.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 2.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 3.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.0 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.5 | GO:0005876 | spindle microtubule(GO:0005876) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 12.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 1.6 | 11.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 1.5 | 11.9 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 1.2 | 4.9 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 1.1 | 4.6 | GO:0004341 | gluconolactonase activity(GO:0004341) |
| 0.9 | 2.6 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.8 | 2.4 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.7 | 7.3 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.7 | 2.0 | GO:0047783 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.7 | 4.0 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.6 | 4.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.6 | 2.8 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.5 | 6.9 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.4 | 4.9 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.4 | 2.2 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.4 | 1.2 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.4 | 1.6 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.3 | 1.5 | GO:0004803 | transposase activity(GO:0004803) |
| 0.3 | 2.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.3 | 3.7 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.3 | 0.8 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.2 | 1.5 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.2 | 4.0 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.2 | 2.8 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.2 | 1.4 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.2 | 2.0 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.2 | 6.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 1.0 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.2 | 0.7 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.2 | 10.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 5.7 | GO:0001848 | complement binding(GO:0001848) |
| 0.2 | 2.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 17.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 0.9 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.2 | 4.8 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 3.6 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 2.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 1.7 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 1.9 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 1.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.8 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 1.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 5.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 5.9 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 9.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 3.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.9 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 3.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 2.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 1.6 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 9.1 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.1 | 2.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.2 | GO:0004750 | ribulose-phosphate 3-epimerase activity(GO:0004750) |
| 0.1 | 0.3 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.1 | 1.6 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.1 | 2.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.9 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 13.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 4.5 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.2 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 6.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.4 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 1.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 3.4 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 3.2 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.6 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 3.3 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 3.3 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.2 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.1 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.0 | 5.6 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 0.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.3 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 1.7 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.0 | 0.8 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 1.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 2.1 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 2.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.4 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 1.4 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 5.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 14.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.4 | 6.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.3 | 12.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 5.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 11.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.2 | 6.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.2 | 4.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 8.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 5.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 2.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 15.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.5 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 17.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 3.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 3.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.0 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 4.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 2.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.9 | PID ENDOTHELIN PATHWAY | Endothelins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.9 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.3 | 3.7 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.3 | 5.7 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.2 | 8.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 6.5 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.2 | 3.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.2 | 2.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.2 | 12.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.2 | 2.8 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 2.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 4.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 2.8 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 3.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 11.1 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 5.8 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 2.0 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 4.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 2.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 4.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 6.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 2.9 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 5.0 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 2.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 2.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 1.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.9 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 4.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.2 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 5.3 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |