Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
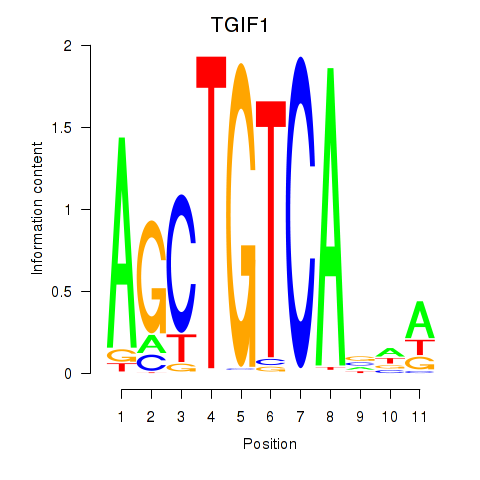
Results for TGIF1
Z-value: 6.68
Transcription factors associated with TGIF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TGIF1
|
ENSG00000177426.22 | TGIF1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TGIF1 | hg38_v1_chr18_+_3451647_3451724 | -0.71 | 4.5e-35 | Click! |
Activity profile of TGIF1 motif
Sorted Z-values of TGIF1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TGIF1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 17.5 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 5.7 | 28.4 | GO:1901628 | positive regulation of postsynaptic membrane organization(GO:1901628) |
| 5.3 | 15.8 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 4.7 | 28.4 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 4.6 | 13.8 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 3.3 | 9.8 | GO:0014873 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 3.2 | 54.4 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 3.2 | 9.5 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 2.9 | 11.6 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 2.7 | 8.2 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 2.6 | 23.8 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 2.4 | 9.6 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 2.2 | 6.6 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 2.1 | 6.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 1.9 | 1.9 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 1.8 | 9.2 | GO:0030070 | insulin processing(GO:0030070) |
| 1.8 | 5.4 | GO:0061590 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 1.7 | 15.4 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 1.7 | 15.4 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 1.7 | 16.9 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 1.5 | 5.9 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 1.4 | 10.1 | GO:0002118 | aggressive behavior(GO:0002118) |
| 1.4 | 4.3 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 1.4 | 13.9 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.4 | 4.1 | GO:0072276 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 1.3 | 22.8 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 1.3 | 11.5 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 1.2 | 2.5 | GO:0003257 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 1.2 | 14.4 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 1.2 | 4.8 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 1.2 | 3.6 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 1.2 | 4.8 | GO:0070295 | renal water absorption(GO:0070295) |
| 1.1 | 15.6 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 1.1 | 18.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 1.1 | 4.2 | GO:0010157 | response to chlorate(GO:0010157) |
| 1.1 | 3.2 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 1.0 | 4.1 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 1.0 | 4.1 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 1.0 | 4.0 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 1.0 | 3.0 | GO:0060366 | subpallium cell proliferation in forebrain(GO:0022012) lateral ganglionic eminence cell proliferation(GO:0022018) lambdoid suture morphogenesis(GO:0060366) sagittal suture morphogenesis(GO:0060367) anterior semicircular canal development(GO:0060873) lateral semicircular canal development(GO:0060875) |
| 1.0 | 3.0 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 1.0 | 3.0 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) positive regulation of somatostatin secretion(GO:0090274) |
| 1.0 | 8.8 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 1.0 | 2.9 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.9 | 2.7 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.9 | 3.5 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.9 | 9.5 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.8 | 3.4 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.8 | 3.2 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.8 | 4.0 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.8 | 8.8 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.8 | 3.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.7 | 2.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.7 | 11.8 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.7 | 4.1 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.7 | 2.7 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.6 | 2.6 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.6 | 5.8 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.6 | 9.4 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.6 | 3.7 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.6 | 4.2 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.6 | 1.8 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.6 | 1.8 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.6 | 4.7 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.6 | 6.4 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.6 | 4.0 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.6 | 7.9 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.6 | 3.3 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.6 | 23.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.5 | 3.8 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.5 | 2.2 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.5 | 16.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.5 | 5.4 | GO:0051126 | negative regulation of actin nucleation(GO:0051126) |
| 0.5 | 3.7 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.5 | 1.6 | GO:0071934 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.5 | 3.7 | GO:0060492 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.5 | 6.2 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.5 | 15.4 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.5 | 1.5 | GO:0070429 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) |
| 0.5 | 2.8 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.5 | 0.9 | GO:0002329 | pre-B cell differentiation(GO:0002329) pre-B cell allelic exclusion(GO:0002331) |
| 0.5 | 1.8 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.5 | 1.8 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.5 | 1.8 | GO:0048627 | myoblast development(GO:0048627) |
| 0.4 | 13.6 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.4 | 6.1 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.4 | 5.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.4 | 2.6 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.4 | 1.7 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.4 | 1.3 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.4 | 1.2 | GO:0033025 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) mast cell proliferation(GO:0070662) |
| 0.4 | 6.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.4 | 1.2 | GO:0070256 | negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.4 | 1.2 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.4 | 4.0 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.4 | 3.5 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.4 | 1.8 | GO:0051935 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.4 | 10.6 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.3 | 3.7 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.3 | 1.3 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.3 | 2.3 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.3 | 5.7 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.3 | 4.7 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.3 | 4.7 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.3 | 2.2 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.3 | 8.3 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.3 | 3.5 | GO:0097070 | ductus arteriosus closure(GO:0097070) |
| 0.3 | 9.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.3 | 2.3 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.3 | 1.9 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.3 | 8.8 | GO:0009081 | branched-chain amino acid metabolic process(GO:0009081) branched-chain amino acid catabolic process(GO:0009083) |
| 0.3 | 1.4 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.3 | 3.1 | GO:0051967 | regulation of short-term neuronal synaptic plasticity(GO:0048172) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.3 | 4.8 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.2 | 4.7 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 2.5 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.2 | 8.9 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.2 | 1.9 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.2 | 8.6 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.2 | 3.8 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.2 | 6.3 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.2 | 3.3 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.2 | 0.2 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 0.2 | 13.0 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.2 | 1.8 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.2 | 1.1 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.2 | 9.1 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.2 | 1.8 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.2 | 1.4 | GO:0036343 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) psychomotor behavior(GO:0036343) motor behavior(GO:0061744) |
| 0.2 | 1.7 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.2 | 4.0 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.2 | 3.5 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.2 | 6.1 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.2 | 2.9 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.2 | 7.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.2 | 11.6 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.2 | 3.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 0.7 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) neuron projection maintenance(GO:1990535) |
| 0.2 | 2.0 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.2 | 0.5 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.2 | 7.9 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.2 | 3.2 | GO:2000178 | Rap protein signal transduction(GO:0032486) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.2 | 1.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.2 | 4.2 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.2 | 2.2 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.2 | 6.1 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.2 | 0.8 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.2 | 2.9 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 41.0 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.1 | 0.6 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 1.1 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.1 | 0.4 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 4.1 | GO:0015800 | acidic amino acid transport(GO:0015800) L-glutamate transport(GO:0015813) |
| 0.1 | 0.7 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.1 | 12.4 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 20.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 2.3 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 11.6 | GO:0032092 | positive regulation of protein binding(GO:0032092) |
| 0.1 | 0.2 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 | 5.2 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 | 1.0 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 2.3 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.1 | 2.7 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.1 | 5.6 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 0.3 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.1 | 2.4 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.1 | 4.2 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 0.8 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.1 | 3.8 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 1.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 1.6 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.1 | 8.3 | GO:0005977 | glycogen metabolic process(GO:0005977) |
| 0.1 | 2.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 2.8 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.1 | 3.8 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 3.8 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.1 | 3.5 | GO:0030317 | sperm motility(GO:0030317) |
| 0.1 | 0.6 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 0.6 | GO:0000101 | sulfur amino acid transport(GO:0000101) L-cystine transport(GO:0015811) |
| 0.1 | 1.6 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 1.4 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 0.9 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 1.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 3.7 | GO:0032210 | regulation of telomere maintenance via telomerase(GO:0032210) |
| 0.1 | 2.0 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.1 | 0.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 5.3 | GO:0051438 | regulation of ubiquitin-protein transferase activity(GO:0051438) |
| 0.1 | 0.2 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.1 | 0.4 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.1 | 2.7 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 2.6 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.1 | 3.3 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.1 | 5.2 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.1 | 6.6 | GO:0007098 | centrosome cycle(GO:0007098) centrosome organization(GO:0051297) |
| 0.1 | 1.5 | GO:0010863 | positive regulation of phospholipase C activity(GO:0010863) |
| 0.0 | 0.5 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 1.9 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 5.8 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.0 | 0.5 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 1.2 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.6 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 1.7 | GO:0015992 | proton transport(GO:0015992) |
| 0.0 | 3.9 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.9 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 1.4 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) |
| 0.0 | 4.3 | GO:0007626 | locomotory behavior(GO:0007626) |
| 0.0 | 2.4 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 1.8 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.0 | 0.3 | GO:0034434 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.0 | 4.0 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 0.3 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.2 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 1.5 | GO:0031960 | response to corticosteroid(GO:0031960) |
| 0.0 | 0.9 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.6 | GO:0044364 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.0 | 0.6 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 54.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 3.1 | 12.4 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 2.7 | 10.6 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 2.5 | 7.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 1.9 | 9.5 | GO:0001652 | granular component(GO:0001652) |
| 1.6 | 8.2 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 1.4 | 15.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 1.1 | 4.4 | GO:0070695 | FHF complex(GO:0070695) |
| 1.1 | 8.8 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 1.0 | 4.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.9 | 5.6 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.9 | 4.6 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.7 | 28.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.7 | 6.6 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.7 | 3.6 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.7 | 2.8 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.7 | 2.7 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.6 | 1.8 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.6 | 11.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.5 | 25.8 | GO:0031430 | M band(GO:0031430) |
| 0.5 | 3.3 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.5 | 3.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.5 | 3.3 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.5 | 22.1 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.4 | 13.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.4 | 3.6 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.4 | 12.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.4 | 1.8 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.4 | 8.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.3 | 19.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.3 | 0.6 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.3 | 3.7 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.3 | 3.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.3 | 3.7 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.3 | 6.1 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.2 | 9.4 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.2 | 26.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.2 | 2.0 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 4.8 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.2 | 4.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 4.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 13.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 3.7 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.2 | 22.4 | GO:0005901 | caveola(GO:0005901) |
| 0.2 | 8.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.2 | 5.2 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.2 | 6.2 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.2 | 3.9 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 12.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.2 | 7.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.2 | 2.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.2 | 1.8 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.2 | 25.8 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.2 | 5.4 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 28.3 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 4.0 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 1.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 4.0 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 4.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 5.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 1.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 4.6 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 3.5 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 1.3 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 3.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 1.9 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 7.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 16.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.3 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.1 | 3.2 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 13.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 4.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 1.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 25.9 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.1 | 3.0 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 6.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 1.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 4.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 1.0 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.9 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 3.2 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 17.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 0.9 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 2.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 9.2 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.0 | 3.8 | GO:0034702 | ion channel complex(GO:0034702) |
| 0.0 | 14.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 3.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 4.2 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 8.5 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 3.6 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 1.4 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 3.5 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.8 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 5.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 3.9 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 2.0 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.4 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 2.0 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 4.9 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 2.6 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.1 | GO:0098794 | postsynapse(GO:0098794) |
| 0.0 | 1.7 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 5.0 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 14.7 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 3.4 | 13.7 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 3.2 | 9.6 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 3.2 | 9.5 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 2.6 | 15.4 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 2.3 | 6.8 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 1.9 | 15.4 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 1.8 | 9.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 1.7 | 51.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 1.7 | 23.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 1.6 | 23.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 1.5 | 8.8 | GO:0070728 | leucine binding(GO:0070728) |
| 1.5 | 11.6 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 1.4 | 9.9 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 1.4 | 4.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 1.4 | 4.1 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 1.2 | 3.6 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 1.0 | 11.4 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 1.0 | 39.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 1.0 | 11.8 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 1.0 | 7.8 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.9 | 3.5 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.9 | 3.5 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.8 | 18.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.8 | 11.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.8 | 6.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.8 | 6.8 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.7 | 12.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.7 | 15.4 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.7 | 2.9 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.7 | 2.8 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.6 | 2.6 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.6 | 3.9 | GO:0016807 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.6 | 16.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.6 | 2.4 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 0.6 | 3.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.6 | 6.4 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.5 | 4.9 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.5 | 1.6 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.5 | 9.7 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.5 | 3.5 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.5 | 16.9 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.5 | 5.7 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.5 | 3.7 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.4 | 16.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.4 | 1.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.4 | 7.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.4 | 3.2 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.4 | 1.8 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.4 | 5.5 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.4 | 10.6 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.4 | 5.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.4 | 6.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.4 | 3.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.4 | 8.9 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.4 | 19.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.3 | 16.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.3 | 3.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.3 | 3.7 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.3 | 4.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.3 | 2.6 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.3 | 17.4 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.3 | 4.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.3 | 3.7 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.3 | 1.2 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.3 | 4.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.3 | 3.8 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.3 | 2.5 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.3 | 3.5 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.3 | 7.6 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.3 | 6.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.3 | 5.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 7.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 3.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.2 | 1.2 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.2 | 3.8 | GO:0019841 | retinol binding(GO:0019841) |
| 0.2 | 4.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 2.7 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 1.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.2 | 1.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 3.6 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.2 | 6.2 | GO:0071949 | FAD binding(GO:0071949) |
| 0.2 | 3.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.2 | 3.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 6.3 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.2 | 3.8 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.2 | 5.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 1.8 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.2 | 15.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.2 | 3.7 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.2 | 0.8 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 2.7 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.2 | 6.9 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.2 | 6.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 3.6 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.2 | 4.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 7.9 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.2 | 0.9 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.2 | 5.7 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.2 | 1.6 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 1.0 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 6.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.2 | 1.7 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.2 | 3.6 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 0.8 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.2 | 2.4 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.1 | 0.7 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 5.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 0.6 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 2.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 5.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 1.8 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.5 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.7 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.1 | 2.0 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.1 | 4.0 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 3.8 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 6.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 1.3 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.1 | 2.7 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 1.8 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 3.1 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.3 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.1 | 3.7 | GO:0070182 | protein phosphatase 2A binding(GO:0051721) DNA polymerase binding(GO:0070182) |
| 0.1 | 27.3 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 1.7 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 1.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 3.0 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 1.6 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 2.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 0.5 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 3.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 12.5 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 0.6 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.1 | 1.9 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 2.6 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 4.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 1.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 6.0 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 1.9 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 4.0 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 5.5 | GO:0008234 | cysteine-type peptidase activity(GO:0008234) |
| 0.1 | 1.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.3 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.5 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.4 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 3.4 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 3.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 4.8 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 1.0 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 2.3 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.5 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.7 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.7 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.8 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 1.4 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 1.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.8 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 1.8 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 2.4 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 3.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 7.4 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 1.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.8 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 1.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 6.4 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 6.9 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 1.6 | GO:0004386 | helicase activity(GO:0004386) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 25.8 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.5 | 14.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.4 | 17.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.4 | 13.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.4 | 14.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.4 | 14.9 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.3 | 17.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.3 | 9.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.3 | 34.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.2 | 15.7 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.2 | 4.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.2 | 4.7 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.2 | 3.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 7.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 8.8 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 6.7 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 9.2 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 4.0 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 5.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 13.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 18.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.6 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 2.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 3.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 1.9 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 3.0 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 1.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 1.4 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 1.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 2.1 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 4.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 3.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 3.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 2.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.9 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.1 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.0 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 3.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.4 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 4.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 16.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 1.2 | 15.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 1.1 | 11.8 | REACTOME OPSINS | Genes involved in Opsins |
| 1.0 | 34.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.9 | 31.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.6 | 13.7 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.5 | 9.6 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.4 | 1.7 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.4 | 3.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.4 | 20.0 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.4 | 15.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.4 | 11.6 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.3 | 6.2 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.3 | 4.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.3 | 4.7 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.3 | 8.9 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.3 | 6.6 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.3 | 10.1 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.2 | 6.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 1.8 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.2 | 7.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 4.2 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.2 | 2.9 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 10.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 3.2 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.2 | 2.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 3.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 2.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 6.4 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 3.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.9 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 5.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 5.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 6.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 5.6 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.1 | 3.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 7.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 3.1 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.1 | 4.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.6 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 4.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 6.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 2.6 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 3.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 4.6 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 6.3 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.1 | 16.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 9.0 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 1.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 1.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.4 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 1.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 1.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 6.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 0.9 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 4.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 4.2 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 1.1 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 1.2 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 4.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 2.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 3.6 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 3.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.8 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 2.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 1.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.3 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |