Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for THRB
Z-value: 4.26
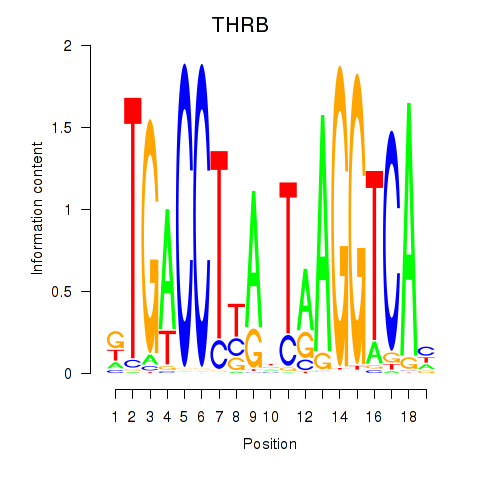
Transcription factors associated with THRB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
THRB
|
ENSG00000151090.20 | THRB |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| THRB | hg38_v1_chr3_-_24494842_24494866, hg38_v1_chr3_-_24165548_24165595, hg38_v1_chr3_-_24494875_24494882 | -0.21 | 2.2e-03 | Click! |
Activity profile of THRB motif
Sorted Z-values of THRB motif
Network of associatons between targets according to the STRING database.
First level regulatory network of THRB
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 24.5 | GO:0090166 | regulation of Schwann cell differentiation(GO:0014038) Golgi disassembly(GO:0090166) |
| 2.6 | 7.8 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 2.4 | 11.8 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 1.9 | 32.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 1.5 | 27.9 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 1.4 | 6.9 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 1.1 | 6.8 | GO:0030581 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.9 | 2.8 | GO:0035744 | T-helper 1 cell cytokine production(GO:0035744) |
| 0.7 | 6.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.6 | 11.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.5 | 3.1 | GO:0016036 | cellular response to phosphate starvation(GO:0016036) positive regulation of sulfur amino acid metabolic process(GO:0031337) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.5 | 2.5 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.4 | 7.9 | GO:0010566 | ubiquinone biosynthetic process(GO:0006744) regulation of ketone biosynthetic process(GO:0010566) quinone biosynthetic process(GO:1901663) |
| 0.3 | 4.0 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 8.4 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 0.3 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 | 0.7 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.2 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 24.5 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 2.8 | 27.9 | GO:0070852 | cell body fiber(GO:0070852) |
| 1.7 | 6.8 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 1.0 | 6.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.3 | 4.0 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.2 | 8.4 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 7.8 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 32.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 11.8 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 13.2 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 2.8 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 3.1 | GO:0043197 | dendritic spine(GO:0043197) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 32.0 | GO:0004459 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 2.6 | 7.9 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 1.9 | 7.8 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 1.1 | 24.5 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 1.0 | 27.9 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.6 | 6.9 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.5 | 6.8 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.5 | 11.8 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.4 | 11.7 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.3 | 3.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.2 | 8.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 0.9 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 0.7 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 6.2 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 4.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.2 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 5.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 1.2 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 24.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 8.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 6.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 24.5 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.9 | 31.1 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 11.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 4.6 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 3.1 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 6.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.7 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |