Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for TLX1_NFIC
Z-value: 4.70
Transcription factors associated with TLX1_NFIC
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
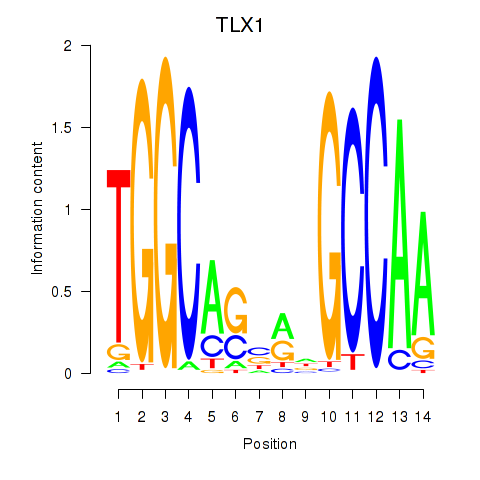
TLX1
|
ENSG00000107807.13 | TLX1 |
|
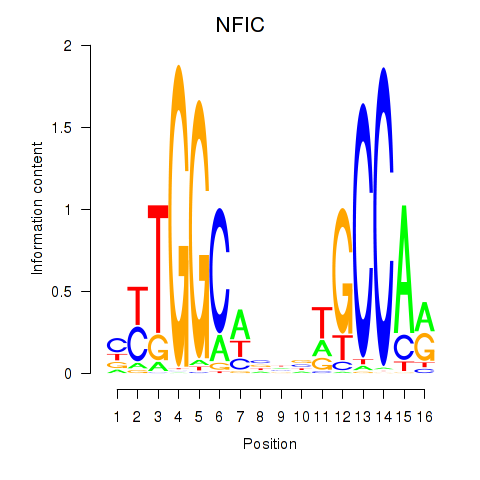
NFIC
|
ENSG00000141905.19 | NFIC |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFIC | hg38_v1_chr19_+_3366549_3366621 | 0.42 | 6.6e-11 | Click! |
| TLX1 | hg38_v1_chr10_+_101131284_101131321 | 0.24 | 4.6e-04 | Click! |
Activity profile of TLX1_NFIC motif
Sorted Z-values of TLX1_NFIC motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TLX1_NFIC
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 20.9 | 104.6 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 13.6 | 67.8 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 13.3 | 53.3 | GO:1902228 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 11.7 | 46.9 | GO:1903631 | regulation of calcium-dependent ATPase activity(GO:1903610) negative regulation of calcium-dependent ATPase activity(GO:1903611) regulation of dUTP diphosphatase activity(GO:1903627) positive regulation of dUTP diphosphatase activity(GO:1903629) negative regulation of aminoacyl-tRNA ligase activity(GO:1903631) regulation of leucine-tRNA ligase activity(GO:1903633) negative regulation of leucine-tRNA ligase activity(GO:1903634) |
| 10.9 | 43.7 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 10.0 | 30.0 | GO:0061027 | umbilical cord morphogenesis(GO:0036304) umbilical cord development(GO:0061027) |
| 6.7 | 20.1 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 6.2 | 24.9 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 5.3 | 89.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 4.7 | 14.2 | GO:0001300 | chronological cell aging(GO:0001300) |
| 4.7 | 14.1 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 4.1 | 49.4 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 3.5 | 10.5 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 3.5 | 13.9 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 3.1 | 21.7 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 3.0 | 9.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 2.8 | 8.5 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 2.8 | 30.6 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 2.8 | 24.8 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 2.7 | 5.3 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 2.6 | 7.9 | GO:0060005 | vestibular reflex(GO:0060005) |
| 2.5 | 45.5 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 2.5 | 17.6 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 2.4 | 49.5 | GO:0032060 | bleb assembly(GO:0032060) |
| 2.3 | 9.2 | GO:0019474 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 2.3 | 6.8 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 2.3 | 20.4 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 2.2 | 8.9 | GO:2000366 | response to prolactin(GO:1990637) regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 2.1 | 10.7 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 2.1 | 18.9 | GO:0009407 | toxin catabolic process(GO:0009407) secondary metabolite catabolic process(GO:0090487) |
| 2.1 | 8.3 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 1.9 | 7.7 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 1.9 | 36.0 | GO:0015671 | oxygen transport(GO:0015671) |
| 1.9 | 7.5 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 1.9 | 9.3 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 1.8 | 37.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 1.8 | 15.8 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 1.7 | 51.5 | GO:0031639 | plasminogen activation(GO:0031639) |
| 1.7 | 6.8 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 1.6 | 26.0 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 1.6 | 28.5 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 1.5 | 4.4 | GO:0006524 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-amino acid metabolic process(GO:0046416) |
| 1.4 | 5.6 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 1.3 | 16.0 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 1.3 | 4.0 | GO:1901205 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) negative regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901205) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 1.2 | 4.9 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 1.2 | 7.3 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 1.2 | 6.1 | GO:0051935 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 1.1 | 11.5 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 1.0 | 10.3 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 1.0 | 7.2 | GO:0045007 | depurination(GO:0045007) |
| 1.0 | 28.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 1.0 | 15.1 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 1.0 | 3.9 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.9 | 16.1 | GO:0038065 | collagen-activated signaling pathway(GO:0038065) |
| 0.9 | 121.2 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.9 | 10.0 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.8 | 3.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.7 | 12.6 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.7 | 13.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.7 | 4.9 | GO:0015705 | iodide transport(GO:0015705) |
| 0.6 | 1.9 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.6 | 5.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.6 | 2.5 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.6 | 1.9 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.6 | 4.4 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.6 | 3.8 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.6 | 4.3 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.6 | 3.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.6 | 9.0 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.6 | 5.7 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.5 | 1.6 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.5 | 11.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.5 | 1.5 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.5 | 5.2 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.5 | 1.4 | GO:0061074 | negative regulation of photoreceptor cell differentiation(GO:0046533) regulation of neural retina development(GO:0061074) regulation of retina development in camera-type eye(GO:1902866) |
| 0.4 | 1.3 | GO:1902462 | mesenchymal stem cell proliferation(GO:0097168) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.4 | 8.5 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.4 | 5.6 | GO:0051873 | innate immune response in mucosa(GO:0002227) killing by host of symbiont cells(GO:0051873) |
| 0.4 | 6.0 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.4 | 5.9 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.4 | 6.0 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) |
| 0.4 | 2.0 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.4 | 6.7 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.4 | 3.5 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.4 | 1.9 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.4 | 7.1 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.4 | 7.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.4 | 2.5 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.4 | 21.7 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.4 | 19.0 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.3 | 5.9 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.3 | 1.0 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.3 | 3.8 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.3 | 4.4 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.3 | 4.0 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.3 | 24.0 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) |
| 0.3 | 5.7 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.3 | 4.3 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.3 | 7.3 | GO:0033622 | integrin activation(GO:0033622) |
| 0.3 | 21.8 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.3 | 12.8 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.3 | 1.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.3 | 2.0 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.3 | 3.1 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.3 | 4.7 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.3 | 7.1 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.3 | 3.0 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.3 | 4.7 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.3 | 4.3 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.3 | 11.8 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.2 | 2.7 | GO:0051964 | netrin-activated signaling pathway(GO:0038007) negative regulation of synapse assembly(GO:0051964) |
| 0.2 | 2.6 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.2 | 0.5 | GO:0071072 | positive regulation of glomerular filtration(GO:0003104) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.2 | 0.7 | GO:1904640 | response to methionine(GO:1904640) |
| 0.2 | 0.5 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.2 | 1.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.2 | 2.4 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.2 | 5.7 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.2 | 1.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.2 | 0.6 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.2 | 2.9 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.2 | 7.6 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.2 | 1.2 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.2 | 1.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 1.3 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.2 | 2.0 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 0.4 | GO:0035712 | T-helper 2 cell activation(GO:0035712) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.2 | 2.0 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.2 | 0.5 | GO:0099557 | trans-synaptic signaling by trans-synaptic complex(GO:0099545) trans-synaptic signaling by trans-synaptic complex, modulating synaptic transmission(GO:0099557) |
| 0.2 | 2.6 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.2 | 5.0 | GO:0021799 | cerebral cortex radially oriented cell migration(GO:0021799) |
| 0.1 | 1.3 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 5.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 4.3 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.1 | 1.8 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 1.8 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.1 | 6.7 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.1 | 12.4 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.1 | 12.0 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 3.7 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 5.2 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.1 | 10.1 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 1.8 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.1 | 4.2 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 3.2 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 41.3 | GO:0051480 | regulation of cytosolic calcium ion concentration(GO:0051480) |
| 0.1 | 3.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 43.7 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.1 | 0.6 | GO:0060282 | negative regulation of cardiac muscle contraction(GO:0055118) positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.9 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 1.4 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.1 | 0.5 | GO:0021648 | vestibulocochlear nerve morphogenesis(GO:0021648) vestibulocochlear nerve formation(GO:0021650) |
| 0.1 | 3.5 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.1 | 2.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 1.0 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.1 | 1.0 | GO:2000463 | positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.1 | 4.3 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 0.8 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.0 | 1.7 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 3.4 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 3.5 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.6 | GO:0003299 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 8.6 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 0.7 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 8.0 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 8.0 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 2.5 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 0.3 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 1.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 1.1 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 7.5 | GO:0048511 | rhythmic process(GO:0048511) |
| 0.0 | 2.5 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 2.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.2 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 0.2 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.3 | 67.8 | GO:0033269 | internode region of axon(GO:0033269) |
| 9.0 | 36.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 8.9 | 53.3 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 8.6 | 129.5 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 7.2 | 21.7 | GO:0070701 | mucus layer(GO:0070701) |
| 5.6 | 89.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 4.7 | 14.2 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 4.3 | 12.8 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 4.0 | 16.1 | GO:0034681 | integrin alpha11-beta1 complex(GO:0034681) |
| 2.6 | 43.7 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 2.3 | 27.0 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 1.7 | 21.8 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 1.5 | 9.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 1.3 | 40.6 | GO:0043218 | compact myelin(GO:0043218) |
| 1.3 | 6.3 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.9 | 8.3 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.9 | 7.3 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.8 | 2.5 | GO:0097180 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.8 | 11.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.8 | 15.8 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.8 | 12.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.7 | 7.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.7 | 2.0 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.5 | 10.1 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.5 | 6.0 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.5 | 49.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.5 | 2.9 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.5 | 0.5 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.5 | 0.9 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.5 | 8.2 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.4 | 13.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.4 | 49.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.4 | 16.3 | GO:0008305 | integrin complex(GO:0008305) |
| 0.3 | 21.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.3 | 22.4 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.3 | 27.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.3 | 4.3 | GO:0033177 | proton-transporting two-sector ATPase complex, proton-transporting domain(GO:0033177) proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.3 | 43.0 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.2 | 13.6 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 32.0 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.2 | 8.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 16.3 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.2 | 9.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.2 | 6.7 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 1.3 | GO:0060091 | kinocilium(GO:0060091) |
| 0.2 | 3.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 16.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 70.6 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.1 | 4.0 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 10.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 4.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 1.9 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 9.4 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 1.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 3.8 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.1 | 33.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 50.7 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 10.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 7.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 4.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 24.4 | GO:0005938 | cell cortex(GO:0005938) |
| 0.1 | 1.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 50.2 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 24.4 | GO:0045121 | membrane raft(GO:0045121) |
| 0.1 | 2.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 3.4 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 11.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.8 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 2.3 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.1 | 2.0 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 8.9 | GO:0030425 | dendrite(GO:0030425) |
| 0.1 | 6.0 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.7 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 1.5 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 3.5 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 1.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 6.3 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 0.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 4.6 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.5 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.2 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 1.4 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 33.2 | 132.8 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) |
| 11.7 | 46.9 | GO:0004341 | gluconolactonase activity(GO:0004341) |
| 10.9 | 43.7 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 10.7 | 53.3 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 9.1 | 36.6 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 7.5 | 30.0 | GO:0035939 | microsatellite binding(GO:0035939) |
| 5.6 | 67.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 5.3 | 15.8 | GO:0035375 | zymogen binding(GO:0035375) |
| 4.6 | 13.9 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 4.5 | 104.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 3.8 | 18.9 | GO:0045174 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 3.6 | 10.7 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 3.5 | 14.1 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 3.5 | 10.5 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) |
| 3.3 | 99.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 3.2 | 12.8 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 2.8 | 8.3 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 2.5 | 17.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 2.4 | 36.0 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 2.3 | 27.0 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 2.2 | 6.7 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 2.2 | 6.6 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 2.0 | 16.1 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 1.8 | 9.2 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 1.8 | 14.2 | GO:0005534 | galactose binding(GO:0005534) |
| 1.6 | 4.9 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 1.6 | 4.8 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 1.5 | 4.6 | GO:0005055 | laminin receptor activity(GO:0005055) |
| 1.4 | 4.3 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 1.4 | 4.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 1.3 | 4.0 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 1.3 | 3.9 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 1.3 | 7.7 | GO:0050051 | leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 1.2 | 13.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 1.2 | 9.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 1.1 | 25.0 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 1.0 | 6.3 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 1.0 | 28.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.9 | 5.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.9 | 12.4 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.9 | 3.5 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.9 | 4.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.8 | 28.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.8 | 2.5 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.8 | 4.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.8 | 2.4 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.7 | 4.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.7 | 8.4 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.7 | 25.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.7 | 6.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.7 | 4.6 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.6 | 1.9 | GO:0098973 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.6 | 8.9 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.6 | 6.3 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.6 | 2.5 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.6 | 1.8 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.6 | 11.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.6 | 7.0 | GO:0008430 | selenium binding(GO:0008430) |
| 0.5 | 8.9 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.5 | 13.2 | GO:0001848 | complement binding(GO:0001848) |
| 0.5 | 10.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.5 | 7.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.4 | 3.5 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.4 | 6.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.4 | 6.2 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.4 | 51.1 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.4 | 11.3 | GO:0071949 | FAD binding(GO:0071949) |
| 0.4 | 9.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.4 | 1.9 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.4 | 8.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.4 | 1.1 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.4 | 3.8 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.4 | 48.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.4 | 1.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.3 | 5.2 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.3 | 4.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.3 | 6.5 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.3 | 3.7 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.3 | 5.8 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.3 | 2.5 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.3 | 0.8 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.3 | 28.4 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.3 | 1.3 | GO:0001093 | TFIIB-class transcription factor binding(GO:0001093) |
| 0.3 | 1.0 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.2 | 12.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.2 | 1.5 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.2 | 21.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 6.7 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 2.0 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 2.0 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.2 | 1.2 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.2 | 4.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 2.7 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 7.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 2.7 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 1.8 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 2.0 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 3.1 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.1 | 0.9 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.1 | 10.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 5.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 1.3 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.2 | GO:0005151 | interleukin-1, Type II receptor binding(GO:0005151) |
| 0.1 | 0.4 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 16.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 3.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 5.7 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.1 | 3.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 13.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.5 | GO:0048407 | superoxide-generating NADPH oxidase activator activity(GO:0016176) platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 9.0 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 0.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 6.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 1.0 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 15.0 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.1 | 7.4 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.1 | 2.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 12.0 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 22.7 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.5 | GO:1901612 | phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 1.5 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 6.2 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 1.1 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.6 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 1.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 2.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.4 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 1.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 8.9 | ST STAT3 PATHWAY | STAT3 Pathway |
| 1.3 | 50.8 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 1.3 | 57.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 1.2 | 58.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.9 | 33.4 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.9 | 1.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.8 | 10.7 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.7 | 31.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.7 | 90.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.5 | 37.5 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.4 | 22.7 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.4 | 5.6 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.4 | 23.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.4 | 12.4 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.3 | 3.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.3 | 33.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.3 | 5.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.3 | 55.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.3 | 57.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.3 | 10.6 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.2 | 61.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 18.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 5.6 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.1 | 7.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 5.3 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 0.5 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 14.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 7.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 1.5 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 4.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 6.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 3.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 2.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 2.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 53.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 1.1 | 27.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.9 | 21.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.9 | 17.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.7 | 106.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.6 | 1.8 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.5 | 4.7 | REACTOME GAB1 SIGNALOSOME | Genes involved in GAB1 signalosome |
| 0.5 | 25.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.5 | 13.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.5 | 15.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.4 | 28.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.4 | 8.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.4 | 13.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.4 | 6.0 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.4 | 5.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.4 | 6.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.3 | 29.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.3 | 6.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.3 | 4.8 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.3 | 5.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.3 | 11.8 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.3 | 4.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.3 | 40.2 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.2 | 16.4 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.2 | 2.7 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.2 | 4.8 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.2 | 19.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 9.4 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.2 | 4.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 4.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 3.9 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.2 | 8.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.2 | 2.8 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.2 | 6.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 12.6 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.2 | 6.8 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.2 | 4.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 2.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 11.0 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.2 | 4.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.2 | 4.2 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.2 | 2.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 7.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 1.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 1.8 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 6.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 9.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 2.5 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 4.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 5.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 2.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.1 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 3.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |