Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for TLX2
Z-value: 2.69
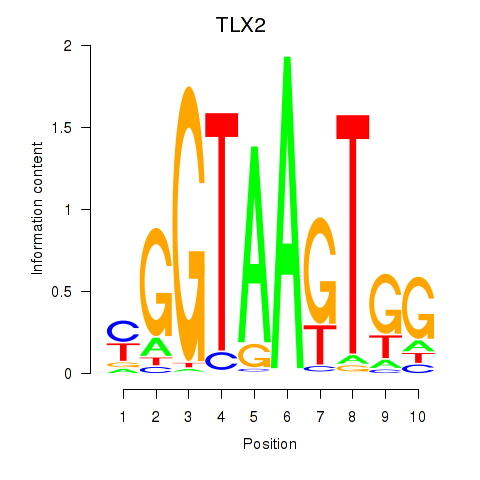
Transcription factors associated with TLX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TLX2
|
ENSG00000115297.11 | TLX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TLX2 | hg38_v1_chr2_+_74514442_74514494, hg38_v1_chr2_+_74513441_74513559 | 0.31 | 4.1e-06 | Click! |
Activity profile of TLX2 motif
Sorted Z-values of TLX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TLX2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 13.5 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 2.9 | 8.8 | GO:0002884 | negative regulation of hypersensitivity(GO:0002884) |
| 2.9 | 11.5 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 2.2 | 13.0 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 1.7 | 5.1 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 1.5 | 5.8 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 1.4 | 9.8 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 1.4 | 4.1 | GO:0014813 | skeletal muscle satellite cell commitment(GO:0014813) |
| 1.2 | 3.6 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 1.2 | 4.7 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 1.1 | 5.5 | GO:0072143 | mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) mesenchyme migration(GO:0090131) |
| 1.1 | 3.2 | GO:1903281 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) positive regulation of potassium ion import(GO:1903288) |
| 0.9 | 2.8 | GO:0051040 | regulation of calcium-independent cell-cell adhesion(GO:0051040) |
| 0.9 | 3.7 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.9 | 4.6 | GO:0035973 | pexophagy(GO:0030242) aggrephagy(GO:0035973) |
| 0.9 | 1.8 | GO:1902263 | induction of programmed cell death(GO:0012502) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.9 | 6.2 | GO:0036018 | response to cobalamin(GO:0033590) cellular response to erythropoietin(GO:0036018) |
| 0.8 | 2.5 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) glomerular visceral epithelial cell migration(GO:0090521) |
| 0.8 | 3.3 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.7 | 2.2 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) negative regulation of membrane invagination(GO:1905154) |
| 0.7 | 2.2 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.7 | 5.7 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.7 | 2.1 | GO:2000584 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.7 | 2.8 | GO:1905069 | hyaluranon cable assembly(GO:0036118) nephrogenic mesenchyme morphogenesis(GO:0072134) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) allantois development(GO:1905069) |
| 0.7 | 9.0 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.7 | 2.1 | GO:0032761 | negative regulation of T cell tolerance induction(GO:0002665) negative regulation of T cell anergy(GO:0002668) negative regulation of lymphocyte anergy(GO:0002912) regulation of lymphotoxin A production(GO:0032681) positive regulation of lymphotoxin A production(GO:0032761) regulation of lymphotoxin A biosynthetic process(GO:0043016) positive regulation of lymphotoxin A biosynthetic process(GO:0043017) |
| 0.7 | 2.1 | GO:0009720 | detection of hormone stimulus(GO:0009720) |
| 0.7 | 2.7 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.6 | 3.2 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.6 | 2.5 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.6 | 1.8 | GO:0021778 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.6 | 4.8 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.6 | 3.0 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.6 | 7.5 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.6 | 2.3 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.6 | 2.8 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.6 | 4.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.5 | 0.5 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.5 | 3.8 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.5 | 1.6 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.5 | 4.3 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.5 | 3.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.5 | 1.5 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.5 | 2.3 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.4 | 4.4 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.4 | 1.7 | GO:0035655 | interleukin-18-mediated signaling pathway(GO:0035655) |
| 0.4 | 3.0 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.4 | 3.8 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.4 | 1.6 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.4 | 2.3 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.4 | 1.1 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.4 | 1.5 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.4 | 2.6 | GO:0032100 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.4 | 1.1 | GO:0051941 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.4 | 1.8 | GO:1900005 | positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.3 | 0.7 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.3 | 4.8 | GO:0070444 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.3 | 1.0 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.3 | 1.0 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.3 | 1.2 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) chemorepulsion of axon(GO:0061643) |
| 0.3 | 3.0 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.3 | 2.7 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.3 | 5.4 | GO:0021794 | thalamus development(GO:0021794) |
| 0.3 | 0.6 | GO:0061054 | dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) positive regulation of dermatome development(GO:0061184) |
| 0.3 | 4.5 | GO:0008212 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.3 | 1.4 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.3 | 2.6 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.3 | 2.0 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.3 | 7.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.2 | 2.5 | GO:0009449 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.2 | 2.7 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 1.4 | GO:1903061 | positive regulation of lipoprotein metabolic process(GO:0050747) regulation of protein lipidation(GO:1903059) positive regulation of protein lipidation(GO:1903061) |
| 0.2 | 2.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.2 | 2.6 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.2 | 1.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.2 | 0.6 | GO:0061386 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 | 2.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.2 | 1.3 | GO:0036102 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.2 | 1.7 | GO:0006477 | protein sulfation(GO:0006477) heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 2.0 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.2 | 2.2 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.2 | 3.4 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.2 | 0.8 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.2 | 1.4 | GO:0036486 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.2 | 2.8 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.2 | 6.0 | GO:0098743 | cell aggregation(GO:0098743) |
| 0.2 | 4.9 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.2 | 1.7 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.2 | 1.7 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.2 | 0.6 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.2 | 7.6 | GO:0014072 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.2 | 1.2 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.2 | 1.4 | GO:0015705 | iodide transport(GO:0015705) |
| 0.2 | 6.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.2 | 0.8 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.2 | 0.7 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.2 | 1.0 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.2 | 0.8 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.2 | 3.9 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.2 | 4.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 2.2 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.2 | 0.5 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.2 | 1.1 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.2 | 1.2 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 1.2 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.1 | 5.5 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.4 | GO:1990654 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 0.1 | 0.6 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.1 | 2.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 4.2 | GO:0097502 | mannosylation(GO:0097502) |
| 0.1 | 1.1 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 | 3.0 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 1.0 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 | 9.4 | GO:0061028 | establishment of endothelial barrier(GO:0061028) |
| 0.1 | 1.0 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.1 | 2.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 3.0 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.1 | 0.7 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 1.4 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) sodium ion export from cell(GO:0036376) |
| 0.1 | 1.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 0.7 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.1 | 0.4 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.1 | 3.0 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 1.4 | GO:0046519 | sphingoid metabolic process(GO:0046519) |
| 0.1 | 8.9 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 1.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 2.1 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.5 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 0.1 | 3.5 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.1 | 0.9 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.1 | 2.8 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 4.0 | GO:1903393 | positive regulation of focal adhesion assembly(GO:0051894) positive regulation of adherens junction organization(GO:1903393) |
| 0.1 | 1.5 | GO:0048485 | sympathetic nervous system development(GO:0048485) |
| 0.1 | 6.6 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 1.3 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 2.7 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 0.8 | GO:0043545 | molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.1 | 2.9 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 1.0 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 0.2 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.1 | 2.8 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.1 | 0.2 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.1 | 1.5 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.1 | 1.2 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 5.7 | GO:0070126 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.1 | 0.5 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 0.4 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.1 | 1.1 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) kinetochore assembly(GO:0051382) |
| 0.1 | 0.8 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.1 | 5.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.6 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 2.3 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.1 | 0.7 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.1 | 0.9 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 4.4 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 3.2 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.1 | 0.8 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.1 | 2.5 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 0.6 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 4.5 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 2.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.8 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 1.4 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 1.3 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.6 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.1 | 1.6 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 9.0 | GO:0030330 | DNA damage response, signal transduction by p53 class mediator(GO:0030330) |
| 0.1 | 0.5 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 4.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 0.5 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 1.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 1.8 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.1 | 0.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 1.3 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 1.7 | GO:0046174 | polyol catabolic process(GO:0046174) |
| 0.0 | 2.8 | GO:0001938 | positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.0 | 0.4 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 1.5 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 | 1.3 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.5 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 1.2 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.2 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 1.2 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.7 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 1.3 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 1.4 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.3 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 1.7 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 1.9 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 1.3 | GO:0030516 | regulation of axon extension(GO:0030516) |
| 0.0 | 1.9 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 0.1 | GO:0070305 | response to cGMP(GO:0070305) |
| 0.0 | 0.5 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.6 | GO:0010666 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.0 | 0.2 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 1.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.3 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 1.9 | GO:0016079 | synaptic vesicle exocytosis(GO:0016079) |
| 0.0 | 0.1 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.0 | 1.8 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 0.7 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 2.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.4 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 1.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 6.5 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 1.2 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 | 1.9 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 1.7 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 2.6 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 6.3 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 1.3 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.8 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 0.2 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.0 | 1.4 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 1.0 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 1.3 | GO:1901264 | carbohydrate derivative transport(GO:1901264) |
| 0.0 | 0.5 | GO:0042311 | vasodilation(GO:0042311) |
| 0.0 | 0.0 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 2.5 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.8 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.4 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.4 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.2 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 3.5 | GO:0090305 | nucleic acid phosphodiester bond hydrolysis(GO:0090305) |
| 0.0 | 0.7 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.7 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 1.9 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 0.8 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.0 | 1.9 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.0 | 1.0 | GO:0070268 | cornification(GO:0070268) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 1.2 | 9.4 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 1.2 | 5.8 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.9 | 6.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.7 | 3.0 | GO:0071546 | pi-body(GO:0071546) |
| 0.7 | 9.0 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.7 | 2.7 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.6 | 4.5 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.5 | 6.1 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.5 | 4.0 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.4 | 8.4 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.4 | 3.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.4 | 8.8 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.4 | 2.3 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.4 | 2.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.4 | 2.5 | GO:1990745 | EARP complex(GO:1990745) |
| 0.4 | 4.9 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.3 | 1.0 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.3 | 3.7 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.3 | 2.7 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.3 | 3.0 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.3 | 3.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 4.4 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.3 | 1.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.3 | 1.7 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.3 | 3.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.3 | 4.1 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.3 | 2.3 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.3 | 2.0 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.3 | 4.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 1.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 3.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.2 | 1.8 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.2 | 1.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.2 | 1.0 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.2 | 6.8 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 0.5 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.2 | 1.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 2.9 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 5.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.2 | 1.8 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 1.5 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 5.4 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 8.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 5.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 2.5 | GO:0044447 | axoneme part(GO:0044447) |
| 0.1 | 0.8 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.1 | 2.1 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.6 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 3.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 1.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 4.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.9 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.1 | 0.4 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 1.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 1.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 13.0 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 5.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 3.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 2.9 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 2.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 2.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.8 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.1 | 1.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.7 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 9.0 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 5.3 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 2.2 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 6.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 2.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 13.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 1.0 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 2.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 2.6 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 4.7 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 0.6 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 19.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 4.6 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 1.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 3.2 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.8 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 5.8 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 2.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 5.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 4.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 1.8 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 2.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 4.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.2 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 2.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 1.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 13.9 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 3.3 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.3 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 1.8 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 1.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 1.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.4 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.7 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 2.8 | 8.4 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 1.9 | 5.7 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 1.6 | 4.9 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 1.1 | 4.5 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 1.1 | 13.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 1.1 | 4.4 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 1.1 | 1.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 1.0 | 3.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 1.0 | 4.8 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.9 | 2.8 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.7 | 2.2 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.7 | 2.2 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.7 | 4.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.7 | 19.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.6 | 3.8 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.6 | 3.0 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.5 | 5.5 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.5 | 2.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.5 | 1.5 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.5 | 7.8 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.5 | 3.4 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.5 | 3.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.5 | 4.7 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.5 | 2.7 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.4 | 1.3 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.4 | 1.7 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.4 | 24.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.4 | 2.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.3 | 4.5 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.3 | 1.4 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.3 | 1.0 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.3 | 1.3 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) scavenger receptor binding(GO:0005124) |
| 0.3 | 9.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.3 | 4.9 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.3 | 0.8 | GO:0034437 | very-low-density lipoprotein particle binding(GO:0034189) glycoprotein transporter activity(GO:0034437) |
| 0.3 | 1.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.3 | 0.8 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.3 | 2.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.3 | 6.1 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.3 | 2.0 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.3 | 3.8 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 1.2 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.2 | 7.5 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 6.7 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.2 | 0.7 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.2 | 3.3 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 2.8 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 1.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 1.7 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.2 | 1.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.2 | 1.1 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.2 | 2.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 1.2 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.2 | 0.5 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.2 | 2.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.2 | 1.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.2 | 3.0 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.2 | 4.2 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.2 | 0.9 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.2 | 0.5 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.2 | 1.2 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 1.2 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 0.4 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.1 | 2.8 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 4.1 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.6 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 1.7 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 1.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 11.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.8 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 3.0 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 2.0 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 1.9 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 1.7 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 1.6 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.9 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 2.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 1.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 2.2 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 0.9 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 4.0 | GO:0015662 | ATPase activity, coupled to transmembrane movement of ions, phosphorylative mechanism(GO:0015662) |
| 0.1 | 0.5 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 3.4 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.1 | 0.7 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 3.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 1.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 5.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.7 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 1.9 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 3.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 2.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.5 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.1 | 1.3 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.1 | 2.8 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 1.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.7 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.5 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.1 | 2.6 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 1.0 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 3.5 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.1 | 1.0 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 0.4 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.1 | 0.6 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 3.9 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 2.5 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 0.9 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 0.9 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 1.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 1.7 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 1.2 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 0.5 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.1 | 0.6 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 1.0 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 1.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.8 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 15.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 6.5 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 7.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.2 | GO:0004341 | gluconolactonase activity(GO:0004341) |
| 0.1 | 1.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.6 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 1.6 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 3.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 2.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.6 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.4 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 1.0 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 1.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 1.0 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 1.0 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.2 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.4 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 4.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 4.2 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 2.4 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 1.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 1.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 9.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.3 | GO:0043813 | phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.0 | 2.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.0 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 2.7 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 1.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.6 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 1.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 2.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.3 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 2.0 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 17.6 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 1.0 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.2 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 10.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.3 | 4.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.3 | 7.8 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.3 | 9.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.3 | 45.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 10.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 3.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 9.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 7.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 4.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 5.7 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 2.8 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 2.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 2.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 3.2 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 1.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 1.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 3.8 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 1.2 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.1 | 3.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 3.7 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 3.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 13.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 4.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 3.4 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 1.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 2.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 1.5 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 0.8 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 2.9 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 2.0 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 1.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 2.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.8 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.0 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.9 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.7 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.6 | PID P53 REGULATION PATHWAY | p53 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.3 | 9.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.3 | 4.5 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.3 | 4.7 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.3 | 3.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.3 | 5.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.2 | 9.7 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.2 | 0.9 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.2 | 4.3 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.2 | 10.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 5.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 5.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 5.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 2.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 3.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 2.8 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 0.5 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 5.2 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.1 | 1.1 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 3.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.5 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 7.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 6.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.6 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.1 | 1.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 2.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 3.0 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 1.8 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 2.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 0.5 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.1 | 3.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 2.1 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 1.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 4.0 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 2.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 2.8 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 2.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 3.8 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.1 | 5.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.2 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 1.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 2.7 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.8 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.9 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.8 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 1.8 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 4.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.9 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 3.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.9 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 1.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.0 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 5.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.2 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 1.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 4.8 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |