Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
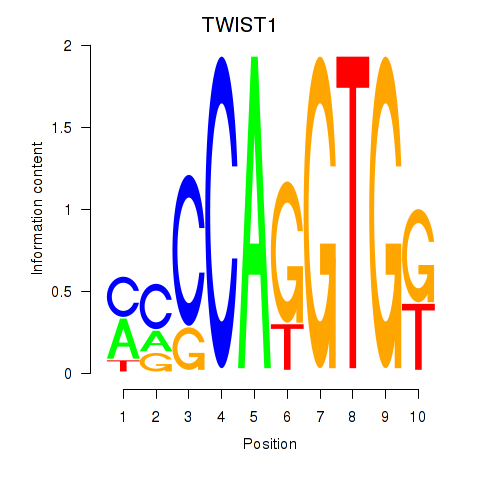
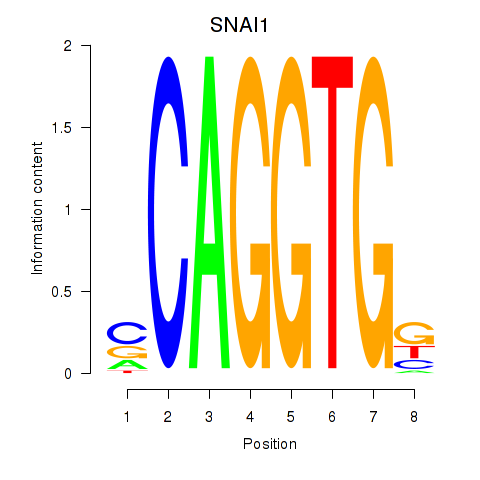
Results for TWIST1_SNAI1
Z-value: 2.13
Transcription factors associated with TWIST1_SNAI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TWIST1
|
ENSG00000122691.13 | TWIST1 |
|
SNAI1
|
ENSG00000124216.4 | SNAI1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TWIST1 | hg38_v1_chr7_-_19117625_19117645 | 0.18 | 6.4e-03 | Click! |
| SNAI1 | hg38_v1_chr20_+_49982969_49982989 | -0.06 | 3.4e-01 | Click! |
Activity profile of TWIST1_SNAI1 motif
Sorted Z-values of TWIST1_SNAI1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TWIST1_SNAI1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.0 | 32.2 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 7.9 | 39.7 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 5.1 | 25.6 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 4.3 | 12.9 | GO:0071283 | regulation of chromosome condensation(GO:0060623) cellular response to iron(III) ion(GO:0071283) |
| 4.1 | 20.3 | GO:0019087 | transformation of host cell by virus(GO:0019087) renal water absorption(GO:0070295) |
| 3.2 | 12.8 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 3.2 | 25.3 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 3.0 | 17.8 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 2.5 | 10.1 | GO:0048627 | myoblast development(GO:0048627) |
| 2.4 | 7.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 2.2 | 6.7 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 2.1 | 10.7 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 2.0 | 7.9 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 2.0 | 9.8 | GO:2000542 | negative regulation of gastrulation(GO:2000542) |
| 1.9 | 5.8 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 1.8 | 3.6 | GO:0002159 | desmosome assembly(GO:0002159) |
| 1.8 | 38.0 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 1.7 | 10.1 | GO:0030581 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 1.6 | 4.9 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 1.5 | 4.5 | GO:0034238 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) |
| 1.4 | 7.2 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 1.4 | 5.5 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 1.4 | 5.5 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 1.4 | 16.3 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 1.4 | 6.8 | GO:0042376 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 1.4 | 56.9 | GO:0097284 | hepatocyte apoptotic process(GO:0097284) |
| 1.3 | 13.4 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 1.3 | 5.3 | GO:0006169 | adenosine salvage(GO:0006169) dATP biosynthetic process(GO:0006175) purine deoxyribonucleoside triphosphate biosynthetic process(GO:0009216) |
| 1.3 | 17.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 1.3 | 7.8 | GO:0007296 | vitellogenesis(GO:0007296) |
| 1.3 | 3.9 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 1.3 | 3.8 | GO:1903892 | negative regulation of ATF6-mediated unfolded protein response(GO:1903892) |
| 1.3 | 3.8 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 1.2 | 17.5 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 1.2 | 4.8 | GO:0033606 | B cell receptor transport within lipid bilayer(GO:0032595) B cell receptor transport into membrane raft(GO:0032597) protein transport out of membrane raft(GO:0032599) chemokine receptor transport out of membrane raft(GO:0032600) negative regulation of transforming growth factor beta3 production(GO:0032913) chemokine receptor transport within lipid bilayer(GO:0033606) |
| 1.2 | 4.6 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 1.1 | 2.2 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 1.1 | 3.3 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 1.1 | 5.5 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 1.1 | 6.6 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 1.1 | 3.3 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 1.1 | 6.3 | GO:0051563 | microglia differentiation(GO:0014004) microglia development(GO:0014005) smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 1.0 | 4.1 | GO:0060620 | negative regulation of triglyceride catabolic process(GO:0010897) regulation of high-density lipoprotein particle clearance(GO:0010982) regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 1.0 | 3.0 | GO:0008615 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 biosynthetic process(GO:0042819) |
| 1.0 | 3.0 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 1.0 | 2.0 | GO:0042823 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 1.0 | 2.9 | GO:0021592 | fourth ventricle development(GO:0021592) third ventricle development(GO:0021678) |
| 0.9 | 3.8 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.9 | 0.9 | GO:2000359 | regulation of binding of sperm to zona pellucida(GO:2000359) |
| 0.9 | 2.7 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.9 | 3.6 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.9 | 7.9 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.9 | 17.4 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.9 | 18.2 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.9 | 7.7 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.8 | 7.0 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.8 | 2.3 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.8 | 3.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.8 | 5.4 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.8 | 2.3 | GO:1902908 | negative regulation of transforming growth factor beta2 production(GO:0032912) negative regulation of hair follicle maturation(GO:0048817) regulation of melanosome transport(GO:1902908) |
| 0.8 | 3.0 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.7 | 3.0 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.7 | 23.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.7 | 5.1 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.7 | 3.6 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.7 | 2.8 | GO:1903382 | neuron intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0036483) regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903381) negative regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903382) |
| 0.7 | 4.9 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.7 | 5.9 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.6 | 1.9 | GO:0031938 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) regulation of chromatin silencing at telomere(GO:0031938) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.6 | 6.3 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.6 | 3.2 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.6 | 10.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.6 | 3.7 | GO:0016321 | female meiosis chromosome segregation(GO:0016321) |
| 0.6 | 1.8 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.6 | 2.4 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.6 | 1.2 | GO:0031077 | post-embryonic camera-type eye development(GO:0031077) |
| 0.6 | 1.8 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.6 | 11.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.6 | 17.8 | GO:0072662 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.6 | 2.3 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.6 | 1.7 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.5 | 1.1 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.5 | 5.8 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.5 | 1.0 | GO:0003257 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.5 | 2.4 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.5 | 3.2 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.5 | 2.7 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.5 | 2.7 | GO:0002760 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.4 | 3.6 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.4 | 2.6 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.4 | 1.3 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.4 | 2.2 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.4 | 5.5 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.4 | 3.3 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.4 | 1.6 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.4 | 2.5 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.4 | 2.4 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.4 | 5.2 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.4 | 3.6 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.4 | 2.7 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.4 | 1.1 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) |
| 0.4 | 4.4 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.3 | 1.0 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.3 | 0.7 | GO:0036486 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.3 | 3.0 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.3 | 3.4 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.3 | 1.3 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.3 | 2.3 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.3 | 2.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.3 | 1.3 | GO:0043387 | toxin catabolic process(GO:0009407) mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) secondary metabolite catabolic process(GO:0090487) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.3 | 3.9 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.3 | 22.1 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.3 | 1.1 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.3 | 0.6 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.3 | 0.8 | GO:0001188 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.3 | 2.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.3 | 0.8 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.3 | 1.6 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.3 | 0.8 | GO:0099541 | trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.3 | 0.8 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.3 | 1.8 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.3 | 6.8 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.3 | 1.8 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.3 | 1.5 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.2 | 1.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 18.2 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.2 | 6.6 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.2 | 3.6 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.2 | 3.1 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.2 | 1.9 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.2 | 0.7 | GO:0016999 | antibiotic metabolic process(GO:0016999) cellular amide catabolic process(GO:0043605) |
| 0.2 | 1.0 | GO:0009439 | cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.2 | 7.8 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.2 | 8.1 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.2 | 10.1 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.2 | 4.7 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.2 | 2.3 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.2 | 0.9 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.2 | 0.2 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.2 | 0.7 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.2 | 0.7 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.2 | 1.8 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.2 | 1.1 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.2 | 2.9 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.2 | 1.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.2 | 1.5 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.2 | 2.0 | GO:0060613 | fat pad development(GO:0060613) |
| 0.2 | 3.0 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.2 | 3.4 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.2 | 6.0 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.2 | 1.9 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 1.5 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.2 | 0.6 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.2 | 6.0 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.2 | 2.0 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.2 | 0.6 | GO:0042427 | serotonin biosynthetic process(GO:0042427) phytoalexin metabolic process(GO:0052314) |
| 0.2 | 0.8 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.2 | 1.2 | GO:0050668 | cellular response to phosphate starvation(GO:0016036) positive regulation of sulfur amino acid metabolic process(GO:0031337) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.2 | 2.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.2 | 1.0 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 1.3 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.2 | 0.6 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.2 | 0.8 | GO:0010193 | response to ozone(GO:0010193) |
| 0.2 | 0.9 | GO:1903412 | response to bile acid(GO:1903412) |
| 0.2 | 0.6 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.2 | 1.3 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.2 | 2.9 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.2 | 7.1 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.2 | 8.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.2 | 6.3 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.2 | 4.1 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.2 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.2 | 0.5 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.2 | 2.7 | GO:0007567 | parturition(GO:0007567) |
| 0.2 | 1.3 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.2 | 2.5 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.2 | 1.5 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.2 | 1.5 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.2 | 0.8 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.2 | 1.6 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.2 | 0.6 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.2 | 0.2 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 0.2 | 2.4 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.2 | 2.9 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.2 | 2.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.2 | 5.7 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.2 | 2.8 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.2 | 1.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.2 | 1.9 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 0.5 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.2 | 3.2 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.1 | 0.6 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.4 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 2.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 1.1 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.1 | 2.0 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 2.4 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.1 | 1.5 | GO:0015868 | purine nucleotide transport(GO:0015865) purine ribonucleotide transport(GO:0015868) |
| 0.1 | 1.0 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.1 | 1.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.5 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 0.4 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) extrathymic T cell selection(GO:0045062) cellular response to interleukin-15(GO:0071350) |
| 0.1 | 3.6 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 1.8 | GO:0036158 | outer dynein arm assembly(GO:0036158) inner dynein arm assembly(GO:0036159) |
| 0.1 | 1.4 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.1 | 1.2 | GO:1900004 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.1 | 0.9 | GO:1905123 | regulation of glucosylceramidase activity(GO:1905123) |
| 0.1 | 5.8 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 1.5 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.4 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.1 | 1.6 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.1 | 1.2 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.1 | 1.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 15.5 | GO:0030509 | BMP signaling pathway(GO:0030509) |
| 0.1 | 0.7 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 2.6 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.1 | 0.4 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.1 | 1.5 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 2.9 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 2.3 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.1 | 1.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 0.7 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.1 | 1.8 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.1 | 0.3 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.1 | 4.7 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 0.7 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 0.8 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.1 | 4.6 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 0.8 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.1 | 1.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.4 | GO:0036114 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.1 | 4.0 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.1 | 0.4 | GO:0061083 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.1 | 5.5 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.1 | 0.5 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 1.1 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 0.7 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 1.1 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.1 | 6.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 1.6 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 2.5 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.1 | 0.3 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.1 | 2.4 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.1 | 0.6 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.1 | 0.3 | GO:2000595 | optic nerve formation(GO:0021634) optic chiasma development(GO:0061360) regulation of optic nerve formation(GO:2000595) positive regulation of optic nerve formation(GO:2000597) |
| 0.1 | 0.3 | GO:0070781 | protein biotinylation(GO:0009305) response to biotin(GO:0070781) histone biotinylation(GO:0071110) |
| 0.1 | 5.6 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.1 | 1.5 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.1 | 1.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.8 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 6.3 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 1.4 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 1.5 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 1.1 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.1 | 9.3 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 1.3 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.1 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 2.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.4 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.1 | 1.6 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.1 | 1.6 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 1.3 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.1 | 0.9 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 1.2 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of adherens junction organization(GO:1903392) |
| 0.1 | 1.6 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 0.7 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.1 | 1.0 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 2.4 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.1 | 1.5 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.9 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.1 | 0.6 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.1 | 3.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 0.4 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 3.9 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 0.4 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.1 | 0.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.2 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 2.0 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 0.6 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 0.4 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.1 | 0.1 | GO:0002331 | pre-B cell differentiation(GO:0002329) pre-B cell allelic exclusion(GO:0002331) |
| 0.1 | 0.3 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.1 | 0.3 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.1 | 0.9 | GO:1903760 | hard palate development(GO:0060022) regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.1 | 0.2 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of epithelial cell differentiation involved in kidney development(GO:2000698) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.1 | 1.6 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 0.8 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.1 | 1.9 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 5.0 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.1 | 0.2 | GO:0015911 | plasma membrane long-chain fatty acid transport(GO:0015911) |
| 0.1 | 1.0 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.6 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.1 | 0.3 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 1.6 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 1.1 | GO:1904376 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.6 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 0.7 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.8 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.7 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 12.6 | GO:0016050 | vesicle organization(GO:0016050) |
| 0.0 | 0.2 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.0 | 2.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.0 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 7.1 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.5 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 1.3 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 1.4 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.8 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 1.2 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 | 1.6 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.3 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.9 | GO:0051084 | 'de novo' posttranslational protein folding(GO:0051084) |
| 0.0 | 0.4 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 1.0 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.5 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.5 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.1 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.0 | 0.7 | GO:1902001 | carnitine shuttle(GO:0006853) fatty acid transmembrane transport(GO:1902001) |
| 0.0 | 0.0 | GO:0072602 | interleukin-4 secretion(GO:0072602) positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.4 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.8 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.4 | GO:0097352 | autophagosome maturation(GO:0097352) vacuole fusion(GO:0097576) |
| 0.0 | 0.6 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 0.3 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.0 | 0.1 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.0 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.2 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.0 | 8.7 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.0 | 0.5 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.0 | 0.5 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.1 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 0.0 | 0.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 2.0 | GO:0007588 | excretion(GO:0007588) |
| 0.0 | 0.8 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.9 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0051106 | positive regulation of DNA ligation(GO:0051106) |
| 0.0 | 1.4 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.0 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.5 | GO:0000305 | response to oxygen radical(GO:0000305) |
| 0.0 | 1.0 | GO:0042472 | inner ear morphogenesis(GO:0042472) |
| 0.0 | 0.2 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.3 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.5 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 3.5 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.2 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 1.5 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.0 | GO:0060003 | copper ion export(GO:0060003) |
| 0.0 | 0.1 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.4 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 0.0 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.1 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.0 | GO:0071442 | regulation of histone H3-K14 acetylation(GO:0071440) positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.5 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.4 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 0.2 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.3 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 48.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 3.3 | 16.4 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 2.4 | 40.9 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 2.3 | 6.9 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 2.3 | 6.8 | GO:0043260 | laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 2.2 | 6.7 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 2.0 | 8.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 1.8 | 14.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 1.7 | 5.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 1.5 | 61.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 1.3 | 6.7 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 1.1 | 3.4 | GO:0070695 | FHF complex(GO:0070695) |
| 1.0 | 32.4 | GO:0030057 | desmosome(GO:0030057) |
| 1.0 | 17.8 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 1.0 | 32.3 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.9 | 11.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.7 | 36.9 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.7 | 14.9 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.7 | 22.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.6 | 1.9 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.6 | 11.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.6 | 26.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.6 | 4.0 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.5 | 3.6 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.5 | 4.6 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.5 | 6.1 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.5 | 4.6 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.5 | 1.5 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.5 | 3.0 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.5 | 1.5 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.5 | 5.0 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.4 | 2.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.4 | 4.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.4 | 0.8 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.4 | 4.0 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.4 | 2.7 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.4 | 1.1 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.3 | 6.9 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.3 | 1.0 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.3 | 2.0 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.3 | 1.8 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.3 | 1.5 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.3 | 0.9 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.3 | 1.1 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.3 | 0.8 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.3 | 2.6 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.3 | 1.8 | GO:0031415 | NatA complex(GO:0031415) |
| 0.3 | 3.0 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.2 | 1.6 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.2 | 1.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 35.2 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.2 | 3.6 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.2 | 2.9 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 2.4 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 1.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.2 | 2.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.2 | 2.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 1.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.2 | 1.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.2 | 59.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.2 | 2.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 3.5 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.2 | 6.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 2.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 2.7 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.2 | 11.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 21.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 5.7 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 1.0 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 2.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 1.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.6 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.1 | 3.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 3.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 4.7 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 0.8 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 2.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 5.5 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 10.1 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 0.7 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 1.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 2.0 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 2.0 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 1.6 | GO:0000800 | lateral element(GO:0000800) |
| 0.1 | 7.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 2.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 8.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 0.7 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 7.0 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 1.5 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 1.8 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 0.6 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 0.4 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.1 | 1.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 0.3 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.1 | 3.0 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 0.3 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 0.6 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 1.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 4.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 1.0 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.1 | 0.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.3 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 2.9 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 11.3 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.5 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 1.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 3.1 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.4 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 3.5 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.8 | GO:0038201 | TORC2 complex(GO:0031932) TOR complex(GO:0038201) |
| 0.0 | 0.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 20.6 | GO:0005925 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.0 | 3.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 4.3 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0033063 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 5.0 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 1.1 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.7 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 1.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 2.1 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 3.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.9 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.1 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.7 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.6 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 3.3 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 1.7 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.0 | 32.2 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 4.1 | 20.3 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 3.5 | 17.4 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 3.2 | 25.3 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 2.7 | 8.1 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 2.6 | 5.2 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 2.5 | 10.1 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 2.5 | 7.5 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 2.4 | 69.0 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 2.2 | 17.7 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 2.2 | 12.9 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 1.9 | 5.8 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 1.8 | 5.5 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 1.8 | 37.0 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 1.3 | 6.7 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 1.3 | 5.3 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 1.2 | 6.2 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 1.1 | 3.2 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 1.1 | 4.3 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 1.0 | 6.2 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 1.0 | 16.3 | GO:0016918 | retinal binding(GO:0016918) |
| 1.0 | 8.8 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 1.0 | 10.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.9 | 3.4 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.8 | 4.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.8 | 6.2 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.8 | 7.0 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.8 | 2.3 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.7 | 2.0 | GO:0031403 | lithium ion binding(GO:0031403) |
| 0.6 | 6.3 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.6 | 17.9 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.6 | 6.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.6 | 2.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.6 | 1.8 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.6 | 1.7 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.6 | 2.8 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.5 | 2.2 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.5 | 9.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.5 | 1.6 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.5 | 4.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.5 | 4.5 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.5 | 47.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.5 | 15.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.5 | 1.4 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.5 | 3.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.4 | 5.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.4 | 5.4 | GO:0008430 | selenium binding(GO:0008430) |
| 0.4 | 4.0 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.4 | 5.5 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.4 | 60.4 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.4 | 7.9 | GO:0043495 | protein anchor(GO:0043495) |
| 0.4 | 2.9 | GO:0004459 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.4 | 5.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.4 | 2.7 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.4 | 1.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.4 | 1.1 | GO:0016420 | [acyl-carrier-protein] S-malonyltransferase activity(GO:0004314) 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) S-malonyltransferase activity(GO:0016419) malonyltransferase activity(GO:0016420) |
| 0.4 | 1.9 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.4 | 2.6 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.4 | 1.1 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.3 | 2.4 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.3 | 5.8 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.3 | 1.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.3 | 4.9 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.3 | 5.5 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.3 | 1.6 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.3 | 1.0 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.3 | 3.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.3 | 1.1 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.3 | 1.7 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.3 | 1.1 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.3 | 1.4 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.3 | 0.8 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.3 | 3.0 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.3 | 3.0 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.3 | 0.8 | GO:0008988 | rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) |
| 0.3 | 0.8 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.3 | 1.5 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.2 | 3.7 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.2 | 2.2 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 1.7 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.2 | 1.3 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.2 | 0.9 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.2 | 3.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 2.7 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.2 | 1.0 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.2 | 2.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 2.4 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.2 | 4.4 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.2 | 0.9 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.2 | 0.7 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.2 | 0.9 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.2 | 3.1 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.2 | 2.7 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.2 | 4.6 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.2 | 3.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 0.8 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.2 | 1.0 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.2 | 0.2 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.2 | 0.8 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.2 | 2.5 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.2 | 3.6 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.2 | 2.5 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.2 | 3.8 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.2 | 4.4 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.2 | 5.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 1.9 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 0.9 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 2.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 9.7 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 2.5 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.1 | 0.7 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.1 | 0.7 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 1.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 7.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 2.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.9 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 1.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 1.8 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 0.5 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.1 | 2.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 3.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 2.2 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 0.5 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.4 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 3.7 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 4.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 1.5 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 2.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.5 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.1 | 0.8 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.5 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 5.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 3.5 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 0.6 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 0.5 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 2.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 1.7 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.3 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 0.4 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 3.5 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 1.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 2.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.7 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.3 | GO:0018271 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 0.1 | 1.6 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 0.2 | GO:0004040 | amidase activity(GO:0004040) |
| 0.1 | 37.1 | GO:0005525 | GTP binding(GO:0005525) |
| 0.1 | 0.6 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.1 | 0.5 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 0.7 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 2.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 3.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.7 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 1.6 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 2.7 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 3.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.2 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.1 | 1.0 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 1.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 4.5 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.1 | 3.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 1.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 1.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 2.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 2.9 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.2 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 0.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.9 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 1.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 1.5 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 0.8 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 1.6 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.1 | 0.6 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 5.9 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 0.5 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 1.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 10.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 1.0 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 10.7 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.1 | 1.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 1.1 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.9 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.5 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 1.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 2.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 2.3 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.7 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 2.0 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.8 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.5 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 4.3 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.3 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.3 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.2 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.0 | 0.2 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.3 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 4.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 4.4 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.7 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.3 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 0.9 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.6 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.2 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.0 | 1.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.4 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 2.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.9 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 1.0 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 1.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.4 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.9 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.7 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 1.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 2.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.6 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.0 | 4.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.2 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.4 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.4 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 2.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 1.2 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.6 | GO:0004402 | histone acetyltransferase activity(GO:0004402) peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 3.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0043008 | U4 snRNA binding(GO:0030621) ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.8 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 2.4 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.1 | GO:0016416 | O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 31.7 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.7 | 2.2 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.6 | 17.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.6 | 25.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.6 | 61.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.5 | 18.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.4 | 3.5 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.4 | 26.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.3 | 17.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.3 | 6.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.3 | 11.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 14.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.3 | 23.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.3 | 3.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.3 | 2.6 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.2 | 9.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.2 | 9.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 9.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 4.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 6.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 4.3 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 38.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 7.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 4.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 24.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 4.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 8.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 1.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 3.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 4.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 2.7 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 1.7 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 2.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 0.9 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 2.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 0.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 1.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 1.0 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 0.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 3.1 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.8 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 2.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 2.0 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 2.3 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.5 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 4.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.0 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.7 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.6 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.4 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.5 | PID P73PATHWAY | p73 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 97.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 1.3 | 27.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 1.2 | 32.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 1.0 | 12.9 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.8 | 19.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.7 | 26.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.7 | 11.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.6 | 14.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.6 | 21.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.5 | 21.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.4 | 20.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.4 | 12.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.4 | 7.1 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.4 | 6.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.3 | 4.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.3 | 7.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.3 | 3.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.3 | 8.3 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.2 | 6.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 15.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.2 | 6.6 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.2 | 0.9 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.2 | 3.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 5.8 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.2 | 4.7 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.2 | 1.3 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.2 | 2.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 4.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.2 | 1.6 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.2 | 3.6 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.2 | 2.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 3.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 4.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 4.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 2.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 3.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 3.9 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 1.9 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 1.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 1.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 1.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 4.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 5.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 2.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 2.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 6.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 2.4 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 6.3 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 0.7 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 1.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 3.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 5.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 2.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 2.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 2.1 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 3.6 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 1.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.9 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.1 | 1.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 2.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 9.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 3.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 1.1 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 2.1 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.9 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.1 | 1.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.0 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.1 | 1.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.0 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 3.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.7 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 4.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.7 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.1 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 2.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 3.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.5 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 4.7 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.4 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 4.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 2.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.6 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.2 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 1.9 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.6 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 3.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.4 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |