Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
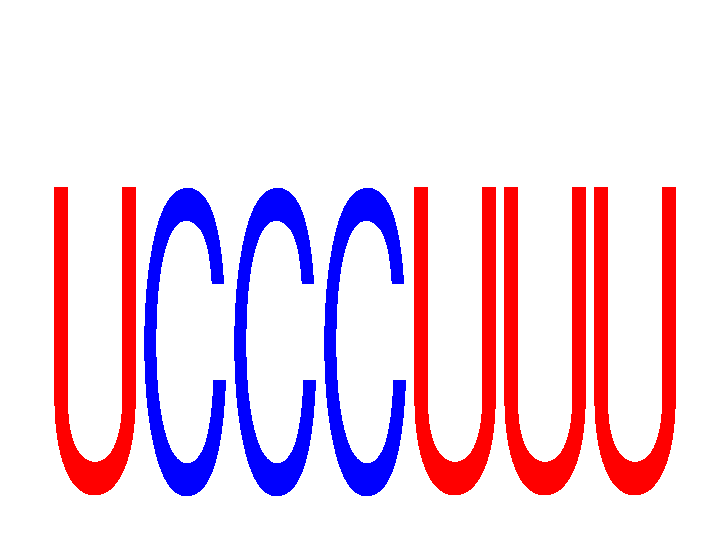
Results for UCCCUUU
Z-value: 1.17
miRNA associated with seed UCCCUUU
| Name | miRBASE accession |
|---|---|
|
hsa-miR-204-5p
|
MIMAT0000265 |
|
hsa-miR-211-5p
|
MIMAT0000268 |
Activity profile of UCCCUUU motif
Sorted Z-values of UCCCUUU motif
Network of associatons between targets according to the STRING database.
First level regulatory network of UCCCUUU
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_108731505 | 1.97 |
ENST00000261401.8
ENST00000552871.5 |
CORO1C
|
coronin 1C |
| chr12_+_65824475 | 1.92 |
ENST00000403681.7
|
HMGA2
|
high mobility group AT-hook 2 |
| chr13_+_97953652 | 1.86 |
ENST00000460070.6
ENST00000481455.6 ENST00000261574.10 ENST00000651721.2 ENST00000493281.6 ENST00000463157.6 ENST00000471898.5 ENST00000489058.6 ENST00000481689.6 |
IPO5
|
importin 5 |
| chr1_-_70205531 | 1.60 |
ENST00000370952.4
|
LRRC40
|
leucine rich repeat containing 40 |
| chr11_-_6683282 | 1.58 |
ENST00000532203.1
ENST00000288937.7 |
MRPL17
|
mitochondrial ribosomal protein L17 |
| chrX_-_20266834 | 1.51 |
ENST00000379565.9
|
RPS6KA3
|
ribosomal protein S6 kinase A3 |
| chr1_-_154970735 | 1.50 |
ENST00000368445.9
ENST00000448116.7 ENST00000368449.8 |
SHC1
|
SHC adaptor protein 1 |
| chr7_+_55887277 | 1.31 |
ENST00000426595.1
|
ENSG00000249773.3
|
novel zinc finger protein 713 (ZNF713) and mitochondrial ribosomal protein S17 (MRPS17) protein |
| chr4_-_110198650 | 1.16 |
ENST00000394607.7
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr7_+_101154445 | 1.10 |
ENST00000337619.11
ENST00000429457.1 |
AP1S1
|
adaptor related protein complex 1 subunit sigma 1 |
| chr3_+_61561561 | 1.09 |
ENST00000474889.6
|
PTPRG
|
protein tyrosine phosphatase receptor type G |
| chrX_+_28587411 | 0.89 |
ENST00000378993.6
|
IL1RAPL1
|
interleukin 1 receptor accessory protein like 1 |
| chr8_-_121641424 | 0.88 |
ENST00000303924.5
|
HAS2
|
hyaluronan synthase 2 |
| chr11_+_63938971 | 0.86 |
ENST00000539656.5
ENST00000377793.9 |
NAA40
|
N-alpha-acetyltransferase 40, NatD catalytic subunit |
| chr15_+_56918612 | 0.84 |
ENST00000438423.6
ENST00000267811.9 ENST00000333725.10 ENST00000559609.5 |
TCF12
|
transcription factor 12 |
| chr19_+_34428353 | 0.83 |
ENST00000590048.6
ENST00000246548.9 |
UBA2
|
ubiquitin like modifier activating enzyme 2 |
| chr6_+_159969070 | 0.81 |
ENST00000356956.6
|
IGF2R
|
insulin like growth factor 2 receptor |
| chr2_-_105329685 | 0.80 |
ENST00000393359.7
|
TGFBRAP1
|
transforming growth factor beta receptor associated protein 1 |
| chr6_-_8102481 | 0.79 |
ENST00000502429.5
ENST00000429723.6 ENST00000379715.10 ENST00000507463.1 |
EEF1E1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr2_+_200811882 | 0.79 |
ENST00000409600.6
|
BZW1
|
basic leucine zipper and W2 domains 1 |
| chr6_-_158818225 | 0.79 |
ENST00000337147.11
|
EZR
|
ezrin |
| chr5_+_95731300 | 0.76 |
ENST00000379982.8
|
RHOBTB3
|
Rho related BTB domain containing 3 |
| chr2_-_199457931 | 0.75 |
ENST00000417098.6
|
SATB2
|
SATB homeobox 2 |
| chr13_-_31161890 | 0.71 |
ENST00000320027.10
|
HSPH1
|
heat shock protein family H (Hsp110) member 1 |
| chr16_-_20900319 | 0.69 |
ENST00000564349.5
ENST00000324344.9 |
ERI2
DCUN1D3
|
ERI1 exoribonuclease family member 2 defective in cullin neddylation 1 domain containing 3 |
| chrX_-_132218124 | 0.67 |
ENST00000342983.6
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr6_+_7107941 | 0.66 |
ENST00000379938.7
ENST00000467782.5 ENST00000334984.10 ENST00000349384.10 |
RREB1
|
ras responsive element binding protein 1 |
| chr9_+_110048598 | 0.66 |
ENST00000434623.6
ENST00000374525.5 |
PALM2AKAP2
|
PALM2 and AKAP2 fusion |
| chr6_-_89352706 | 0.64 |
ENST00000435041.3
|
UBE2J1
|
ubiquitin conjugating enzyme E2 J1 |
| chr3_+_5187697 | 0.64 |
ENST00000256497.9
|
EDEM1
|
ER degradation enhancing alpha-mannosidase like protein 1 |
| chr6_-_85642922 | 0.64 |
ENST00000616122.5
ENST00000678618.1 ENST00000678816.1 ENST00000676630.1 ENST00000678589.1 ENST00000678878.1 ENST00000676542.1 ENST00000355238.11 ENST00000678899.1 |
SYNCRIP
|
synaptotagmin binding cytoplasmic RNA interacting protein |
| chr12_+_4321197 | 0.62 |
ENST00000179259.6
|
TIGAR
|
TP53 induced glycolysis regulatory phosphatase |
| chr10_+_58385395 | 0.61 |
ENST00000487519.6
ENST00000373895.7 |
TFAM
|
transcription factor A, mitochondrial |
| chr1_-_38859669 | 0.61 |
ENST00000373001.4
|
RRAGC
|
Ras related GTP binding C |
| chr20_+_58309704 | 0.59 |
ENST00000244040.4
|
RAB22A
|
RAB22A, member RAS oncogene family |
| chr6_+_15246054 | 0.59 |
ENST00000341776.7
|
JARID2
|
jumonji and AT-rich interaction domain containing 2 |
| chr17_-_68291116 | 0.59 |
ENST00000327268.8
ENST00000580666.6 |
SLC16A6
|
solute carrier family 16 member 6 |
| chr2_+_207529892 | 0.58 |
ENST00000432329.6
ENST00000445803.5 |
CREB1
|
cAMP responsive element binding protein 1 |
| chr6_-_52577012 | 0.55 |
ENST00000182527.4
|
TRAM2
|
translocation associated membrane protein 2 |
| chr7_-_128343823 | 0.55 |
ENST00000415472.6
ENST00000478061.5 ENST00000223073.6 ENST00000459726.1 |
RBM28
|
RNA binding motif protein 28 |
| chr7_+_138460238 | 0.54 |
ENST00000343526.9
|
TRIM24
|
tripartite motif containing 24 |
| chr5_-_128538230 | 0.52 |
ENST00000262464.9
|
FBN2
|
fibrillin 2 |
| chr9_+_109780292 | 0.50 |
ENST00000374530.7
|
PALM2AKAP2
|
PALM2 and AKAP2 fusion |
| chr3_+_155870623 | 0.50 |
ENST00000295920.7
ENST00000496455.7 |
GMPS
|
guanine monophosphate synthase |
| chr15_+_41231219 | 0.50 |
ENST00000334660.10
ENST00000560397.5 |
CHP1
|
calcineurin like EF-hand protein 1 |
| chr4_+_98995709 | 0.48 |
ENST00000296411.11
ENST00000625963.1 |
METAP1
|
methionyl aminopeptidase 1 |
| chr17_+_57085092 | 0.47 |
ENST00000575322.1
ENST00000337714.8 |
AKAP1
|
A-kinase anchoring protein 1 |
| chr3_+_152299392 | 0.46 |
ENST00000498502.5
ENST00000545754.5 ENST00000357472.7 ENST00000324196.9 |
MBNL1
|
muscleblind like splicing regulator 1 |
| chr2_-_151828408 | 0.46 |
ENST00000295087.13
|
ARL5A
|
ADP ribosylation factor like GTPase 5A |
| chrX_+_71533095 | 0.46 |
ENST00000373719.8
ENST00000373701.7 |
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr4_-_65670339 | 0.46 |
ENST00000273854.7
|
EPHA5
|
EPH receptor A5 |
| chr16_+_68085552 | 0.45 |
ENST00000329524.8
|
NFATC3
|
nuclear factor of activated T cells 3 |
| chr3_+_30606574 | 0.45 |
ENST00000295754.10
ENST00000359013.4 |
TGFBR2
|
transforming growth factor beta receptor 2 |
| chr3_-_190120881 | 0.43 |
ENST00000319332.10
|
P3H2
|
prolyl 3-hydroxylase 2 |
| chr3_+_33114007 | 0.43 |
ENST00000320954.11
|
CRTAP
|
cartilage associated protein |
| chr1_+_193121950 | 0.42 |
ENST00000367435.5
|
CDC73
|
cell division cycle 73 |
| chr5_-_143403611 | 0.41 |
ENST00000394464.7
ENST00000231509.7 |
NR3C1
|
nuclear receptor subfamily 3 group C member 1 |
| chr8_-_56211257 | 0.41 |
ENST00000316981.8
ENST00000423799.6 ENST00000429357.2 |
PLAG1
|
PLAG1 zinc finger |
| chr1_+_65147514 | 0.41 |
ENST00000545314.5
|
AK4
|
adenylate kinase 4 |
| chr11_-_57515686 | 0.40 |
ENST00000533263.1
ENST00000278426.8 |
SLC43A1
|
solute carrier family 43 member 1 |
| chr6_+_21593742 | 0.39 |
ENST00000244745.4
|
SOX4
|
SRY-box transcription factor 4 |
| chr16_-_73048104 | 0.38 |
ENST00000268489.10
|
ZFHX3
|
zinc finger homeobox 3 |
| chr7_-_32490361 | 0.37 |
ENST00000410044.5
ENST00000450169.7 ENST00000409987.5 ENST00000409782.5 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr7_+_107580215 | 0.37 |
ENST00000465919.5
ENST00000005259.9 ENST00000445771.6 ENST00000479917.5 ENST00000421217.5 ENST00000457837.5 |
BCAP29
|
B cell receptor associated protein 29 |
| chr6_+_10555787 | 0.37 |
ENST00000316170.9
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2 (I blood group) |
| chr11_+_34051722 | 0.36 |
ENST00000341394.9
ENST00000389645.7 |
CAPRIN1
|
cell cycle associated protein 1 |
| chr11_+_114400592 | 0.33 |
ENST00000541475.5
|
RBM7
|
RNA binding motif protein 7 |
| chr1_+_89821003 | 0.32 |
ENST00000525774.5
ENST00000337338.9 |
LRRC8D
|
leucine rich repeat containing 8 VRAC subunit D |
| chr3_+_170222412 | 0.31 |
ENST00000295797.5
|
PRKCI
|
protein kinase C iota |
| chr7_+_94656325 | 0.31 |
ENST00000482108.1
ENST00000488574.5 ENST00000612748.1 ENST00000613043.1 |
PEG10
|
paternally expressed 10 |
| chr3_+_132417487 | 0.30 |
ENST00000260818.11
|
DNAJC13
|
DnaJ heat shock protein family (Hsp40) member C13 |
| chr8_-_100952918 | 0.30 |
ENST00000395957.6
ENST00000395948.6 ENST00000457309.2 |
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta |
| chr10_+_86756580 | 0.29 |
ENST00000372037.8
|
BMPR1A
|
bone morphogenetic protein receptor type 1A |
| chr11_-_27472698 | 0.29 |
ENST00000389858.4
ENST00000379214.9 |
LGR4
|
leucine rich repeat containing G protein-coupled receptor 4 |
| chr15_+_51829644 | 0.29 |
ENST00000308580.12
|
TMOD3
|
tropomodulin 3 |
| chr2_-_229921963 | 0.28 |
ENST00000343290.5
ENST00000389044.8 ENST00000675453.1 ENST00000675903.1 ENST00000283943.9 |
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr3_-_100401028 | 0.27 |
ENST00000284320.6
|
TOMM70
|
translocase of outer mitochondrial membrane 70 |
| chr5_+_65926556 | 0.27 |
ENST00000380943.6
ENST00000416865.6 ENST00000380935.5 ENST00000284037.10 |
ERBIN
|
erbb2 interacting protein |
| chr14_+_57268963 | 0.26 |
ENST00000261558.8
|
AP5M1
|
adaptor related protein complex 5 subunit mu 1 |
| chr4_-_67701113 | 0.26 |
ENST00000420827.2
ENST00000322244.10 |
UBA6
|
ubiquitin like modifier activating enzyme 6 |
| chr10_+_70815889 | 0.25 |
ENST00000373202.8
|
SGPL1
|
sphingosine-1-phosphate lyase 1 |
| chr22_-_41621014 | 0.25 |
ENST00000263256.7
|
DESI1
|
desumoylating isopeptidase 1 |
| chr17_-_81927699 | 0.24 |
ENST00000574686.1
ENST00000357736.9 |
MAFG
|
MAF bZIP transcription factor G |
| chrX_-_120560947 | 0.24 |
ENST00000674137.11
ENST00000371322.11 ENST00000681090.1 |
CUL4B
|
cullin 4B |
| chr15_-_74938027 | 0.23 |
ENST00000564811.1
ENST00000562233.5 ENST00000322347.11 ENST00000567270.5 ENST00000568783.5 |
COX5A
|
cytochrome c oxidase subunit 5A |
| chr10_-_119596495 | 0.23 |
ENST00000369093.6
|
TIAL1
|
TIA1 cytotoxic granule associated RNA binding protein like 1 |
| chr19_-_14518383 | 0.23 |
ENST00000254322.3
ENST00000595139.2 |
DNAJB1
|
DnaJ heat shock protein family (Hsp40) member B1 |
| chr2_-_131093378 | 0.23 |
ENST00000409185.5
ENST00000389915.4 |
FAM168B
|
family with sequence similarity 168 member B |
| chr4_-_88284553 | 0.22 |
ENST00000608933.6
ENST00000295908.11 |
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent 1K |
| chr13_-_74133892 | 0.22 |
ENST00000377669.7
|
KLF12
|
Kruppel like factor 12 |
| chr13_+_49997019 | 0.21 |
ENST00000420995.6
ENST00000356017.8 ENST00000378182.4 ENST00000457662.2 |
TRIM13
|
tripartite motif containing 13 |
| chr3_-_138834752 | 0.21 |
ENST00000477593.5
ENST00000483968.5 |
PIK3CB
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta |
| chr2_-_171160833 | 0.21 |
ENST00000360843.7
ENST00000431350.7 |
TLK1
|
tousled like kinase 1 |
| chr18_-_63319987 | 0.21 |
ENST00000398117.1
|
BCL2
|
BCL2 apoptosis regulator |
| chr1_+_32013848 | 0.20 |
ENST00000327300.12
ENST00000492989.1 |
KHDRBS1
|
KH RNA binding domain containing, signal transduction associated 1 |
| chrX_+_68499021 | 0.20 |
ENST00000462683.6
|
YIPF6
|
Yip1 domain family member 6 |
| chr22_+_40177917 | 0.19 |
ENST00000454349.7
ENST00000335727.13 |
TNRC6B
|
trinucleotide repeat containing adaptor 6B |
| chr1_+_35883189 | 0.19 |
ENST00000674304.1
ENST00000373204.6 ENST00000674426.1 |
AGO1
|
argonaute RISC component 1 |
| chr10_-_77926724 | 0.18 |
ENST00000372391.7
|
DLG5
|
discs large MAGUK scaffold protein 5 |
| chrX_+_49922605 | 0.18 |
ENST00000376088.7
|
CLCN5
|
chloride voltage-gated channel 5 |
| chr21_-_14383125 | 0.18 |
ENST00000285667.4
|
HSPA13
|
heat shock protein family A (Hsp70) member 13 |
| chr1_+_147541491 | 0.17 |
ENST00000683836.1
ENST00000234739.8 |
BCL9
|
BCL9 transcription coactivator |
| chr12_+_20368495 | 0.17 |
ENST00000359062.4
|
PDE3A
|
phosphodiesterase 3A |
| chr10_+_91923762 | 0.17 |
ENST00000265990.11
|
BTAF1
|
B-TFIID TATA-box binding protein associated factor 1 |
| chr12_-_89352487 | 0.16 |
ENST00000548755.1
ENST00000279488.8 |
DUSP6
|
dual specificity phosphatase 6 |
| chr2_+_96266211 | 0.16 |
ENST00000488633.2
|
CIAO1
|
cytosolic iron-sulfur assembly component 1 |
| chr2_+_5692357 | 0.16 |
ENST00000322002.5
|
SOX11
|
SRY-box transcription factor 11 |
| chr3_+_10141763 | 0.15 |
ENST00000256474.3
ENST00000345392.2 |
VHL
|
von Hippel-Lindau tumor suppressor |
| chrX_-_15854791 | 0.14 |
ENST00000545766.7
ENST00000380291.5 ENST00000672987.1 ENST00000329235.6 |
AP1S2
|
adaptor related protein complex 1 subunit sigma 2 |
| chr2_+_108719473 | 0.14 |
ENST00000283195.11
|
RANBP2
|
RAN binding protein 2 |
| chr18_+_52340179 | 0.13 |
ENST00000442544.7
|
DCC
|
DCC netrin 1 receptor |
| chr2_+_191678122 | 0.13 |
ENST00000425611.9
ENST00000410026.7 |
NABP1
|
nucleic acid binding protein 1 |
| chr7_-_129952901 | 0.13 |
ENST00000472396.5
ENST00000355621.8 |
UBE2H
|
ubiquitin conjugating enzyme E2 H |
| chr17_+_30378903 | 0.13 |
ENST00000225719.9
|
CPD
|
carboxypeptidase D |
| chr12_-_48788995 | 0.13 |
ENST00000550422.5
ENST00000357869.8 |
ADCY6
|
adenylate cyclase 6 |
| chr16_-_71724700 | 0.13 |
ENST00000568954.5
|
PHLPP2
|
PH domain and leucine rich repeat protein phosphatase 2 |
| chr8_+_103819244 | 0.13 |
ENST00000262231.14
ENST00000507740.5 ENST00000408894.6 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr6_+_50818857 | 0.12 |
ENST00000393655.4
|
TFAP2B
|
transcription factor AP-2 beta |
| chr3_-_171460368 | 0.12 |
ENST00000436636.7
ENST00000465393.1 ENST00000341852.10 |
TNIK
|
TRAF2 and NCK interacting kinase |
| chr3_+_184155310 | 0.12 |
ENST00000313143.9
|
DVL3
|
dishevelled segment polarity protein 3 |
| chr3_-_52278620 | 0.12 |
ENST00000296490.8
|
WDR82
|
WD repeat domain 82 |
| chr16_-_30787169 | 0.11 |
ENST00000262525.6
|
ZNF629
|
zinc finger protein 629 |
| chrX_-_112082776 | 0.11 |
ENST00000262839.3
|
TRPC5
|
transient receptor potential cation channel subfamily C member 5 |
| chr5_-_16936231 | 0.11 |
ENST00000507288.1
ENST00000274203.13 ENST00000513610.6 |
MYO10
|
myosin X |
| chr12_-_56636318 | 0.11 |
ENST00000549506.5
ENST00000379441.7 ENST00000551812.5 |
BAZ2A
|
bromodomain adjacent to zinc finger domain 2A |
| chr17_+_32142454 | 0.11 |
ENST00000333942.10
ENST00000358365.7 ENST00000545287.7 |
RHOT1
|
ras homolog family member T1 |
| chr4_-_23890035 | 0.11 |
ENST00000507380.1
ENST00000264867.7 |
PPARGC1A
|
PPARG coactivator 1 alpha |
| chr6_-_16761447 | 0.11 |
ENST00000244769.8
ENST00000436367.6 |
ATXN1
|
ataxin 1 |
| chr5_-_67196791 | 0.11 |
ENST00000256447.5
|
CD180
|
CD180 molecule |
| chr7_-_81770039 | 0.10 |
ENST00000222390.11
ENST00000453411.6 ENST00000457544.7 ENST00000444829.7 |
HGF
|
hepatocyte growth factor |
| chr1_-_22143088 | 0.10 |
ENST00000290167.11
|
WNT4
|
Wnt family member 4 |
| chr1_+_78004930 | 0.09 |
ENST00000370763.6
|
DNAJB4
|
DnaJ heat shock protein family (Hsp40) member B4 |
| chr1_+_2228310 | 0.09 |
ENST00000378536.5
|
SKI
|
SKI proto-oncogene |
| chr8_-_17413345 | 0.09 |
ENST00000180173.10
|
MTMR7
|
myotubularin related protein 7 |
| chr7_+_28412511 | 0.08 |
ENST00000357727.7
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr4_-_139177185 | 0.08 |
ENST00000394235.6
|
ELF2
|
E74 like ETS transcription factor 2 |
| chr7_-_127585566 | 0.08 |
ENST00000321407.3
|
GCC1
|
GRIP and coiled-coil domain containing 1 |
| chr4_-_41748713 | 0.08 |
ENST00000226382.4
|
PHOX2B
|
paired like homeobox 2B |
| chr11_+_112086853 | 0.07 |
ENST00000528182.5
ENST00000528048.5 ENST00000375549.8 ENST00000528021.6 ENST00000526592.5 ENST00000525291.5 |
SDHD
|
succinate dehydrogenase complex subunit D |
| chr1_+_154405193 | 0.07 |
ENST00000622330.4
ENST00000344086.8 |
IL6R
|
interleukin 6 receptor |
| chr7_+_39950187 | 0.06 |
ENST00000181839.10
|
CDK13
|
cyclin dependent kinase 13 |
| chr8_-_42896677 | 0.06 |
ENST00000526349.5
ENST00000534961.5 |
RNF170
|
ring finger protein 170 |
| chr13_-_41961078 | 0.06 |
ENST00000379310.8
ENST00000281496.6 |
VWA8
|
von Willebrand factor A domain containing 8 |
| chr3_+_11272413 | 0.06 |
ENST00000446450.6
ENST00000354956.9 ENST00000354449.7 ENST00000419112.5 |
ATG7
|
autophagy related 7 |
| chr2_-_110115811 | 0.06 |
ENST00000272462.3
|
MALL
|
mal, T cell differentiation protein like |
| chr11_+_35618450 | 0.06 |
ENST00000317811.6
|
FJX1
|
four-jointed box kinase 1 |
| chr1_-_209806124 | 0.05 |
ENST00000367021.8
ENST00000542854.5 |
IRF6
|
interferon regulatory factor 6 |
| chr11_+_35662739 | 0.05 |
ENST00000299413.7
|
TRIM44
|
tripartite motif containing 44 |
| chr10_-_59906509 | 0.05 |
ENST00000263102.7
|
CCDC6
|
coiled-coil domain containing 6 |
| chr11_+_118572373 | 0.04 |
ENST00000392859.7
ENST00000264028.5 ENST00000614498.4 ENST00000359415.8 ENST00000534182.2 |
ARCN1
|
archain 1 |
| chr5_-_37840035 | 0.04 |
ENST00000326524.7
|
GDNF
|
glial cell derived neurotrophic factor |
| chrX_-_24672654 | 0.04 |
ENST00000379145.5
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr1_+_16367088 | 0.04 |
ENST00000471507.5
ENST00000401089.3 ENST00000401088.9 ENST00000492354.1 |
SZRD1
|
SUZ RNA binding domain containing 1 |
| chr10_-_80205551 | 0.03 |
ENST00000372231.7
ENST00000438331.5 |
ANXA11
|
annexin A11 |
| chr17_+_58755821 | 0.03 |
ENST00000308249.4
|
PPM1E
|
protein phosphatase, Mg2+/Mn2+ dependent 1E |
| chr16_-_88941198 | 0.02 |
ENST00000327483.9
ENST00000564416.1 |
CBFA2T3
|
CBFA2/RUNX1 partner transcriptional co-repressor 3 |
| chr9_+_74497308 | 0.02 |
ENST00000376896.8
|
RORB
|
RAR related orphan receptor B |
| chr22_+_32043253 | 0.02 |
ENST00000266088.9
|
SLC5A1
|
solute carrier family 5 member 1 |
| chr12_-_122526929 | 0.01 |
ENST00000331738.12
ENST00000528279.1 ENST00000344591.8 ENST00000526560.6 |
RSRC2
|
arginine and serine rich coiled-coil 2 |
| chr15_-_26773022 | 0.01 |
ENST00000311550.10
ENST00000622697.4 |
GABRB3
|
gamma-aminobutyric acid type A receptor subunit beta3 |
| chr18_+_33578213 | 0.00 |
ENST00000681521.1
ENST00000269197.12 |
ASXL3
|
ASXL transcriptional regulator 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.2 | 0.7 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.2 | 1.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.2 | 0.6 | GO:0036510 | trimming of terminal mannose on C branch(GO:0036510) |
| 0.2 | 0.6 | GO:0043465 | fermentation(GO:0006113) regulation of fermentation(GO:0043465) |
| 0.2 | 0.6 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.2 | 0.7 | GO:0003186 | tricuspid valve development(GO:0003175) tricuspid valve morphogenesis(GO:0003186) |
| 0.2 | 0.9 | GO:1900127 | renal water absorption(GO:0070295) positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.2 | 2.0 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.2 | 0.8 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.6 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.1 | 1.2 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.4 | GO:1901874 | negative regulation of post-translational protein modification(GO:1901874) |
| 0.1 | 0.4 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.1 | 0.6 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.1 | 1.9 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.1 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.1 | 0.7 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.1 | 0.5 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 0.4 | GO:0035910 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.1 | 0.3 | GO:0072204 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.1 | 0.9 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.6 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 0.5 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 0.4 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.3 | GO:1902954 | regulation of early endosome to recycling endosome transport(GO:1902954) |
| 0.1 | 0.5 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.5 | GO:0070885 | positive regulation of sodium:proton antiporter activity(GO:0032417) negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.1 | 0.5 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.1 | 0.5 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 | 0.4 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.7 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.1 | 0.7 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.1 | 0.3 | GO:0070433 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.1 | 0.5 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 0.4 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 0.2 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 0.2 | GO:0046671 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.1 | 0.3 | GO:0021764 | amygdala development(GO:0021764) |
| 0.1 | 0.2 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.6 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.4 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 1.5 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.0 | 0.6 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.4 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.9 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.7 | GO:0045116 | response to UV-C(GO:0010225) protein neddylation(GO:0045116) |
| 0.0 | 1.6 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.2 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.2 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.1 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.0 | 0.2 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.3 | GO:1902915 | negative regulation of histone ubiquitination(GO:0033183) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.1 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.1 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.0 | 0.1 | GO:1904116 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.0 | 0.1 | GO:0061183 | dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) positive regulation of dermatome development(GO:0061184) positive regulation of steroid hormone biosynthetic process(GO:0090031) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.2 | GO:0090625 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.7 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.2 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 1.0 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.0 | 0.1 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.0 | 0.4 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.2 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.3 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.3 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.1 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.0 | 0.1 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.6 | GO:1903432 | regulation of TORC1 signaling(GO:1903432) |
| 0.0 | 0.5 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 1.9 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.0 | GO:0072106 | regulation of ureteric bud formation(GO:0072106) positive regulation of ureteric bud formation(GO:0072107) |
| 0.0 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.4 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.2 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0048880 | sensory system development(GO:0048880) |
| 0.0 | 0.1 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.1 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.2 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.0 | 0.7 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 0.2 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.2 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.2 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.1 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.0 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.3 | 1.9 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.3 | 0.8 | GO:0044393 | microspike(GO:0044393) |
| 0.2 | 0.8 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.2 | 0.8 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.2 | 1.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.8 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 2.0 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 0.6 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 0.5 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.6 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.4 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.6 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 0.4 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.0 | 0.8 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 1.6 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.4 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.9 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.0 | 1.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.3 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.1 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 1.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.6 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.3 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.9 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.3 | 0.9 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.3 | 0.8 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.2 | 1.5 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.2 | 0.9 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.2 | 0.6 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.2 | 0.5 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.2 | 0.5 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.2 | 0.6 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.1 | 0.4 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 1.2 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.4 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.1 | 0.6 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.1 | 1.1 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 0.4 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 0.4 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.1 | 0.6 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 0.5 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 1.9 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 0.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.2 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.1 | 0.9 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.1 | 0.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.5 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.4 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 1.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.3 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 1.1 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.5 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.5 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 1.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 2.1 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.1 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.0 | 1.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.3 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.6 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.8 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 1.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.5 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 2.2 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 0.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.9 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.3 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.9 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |