Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
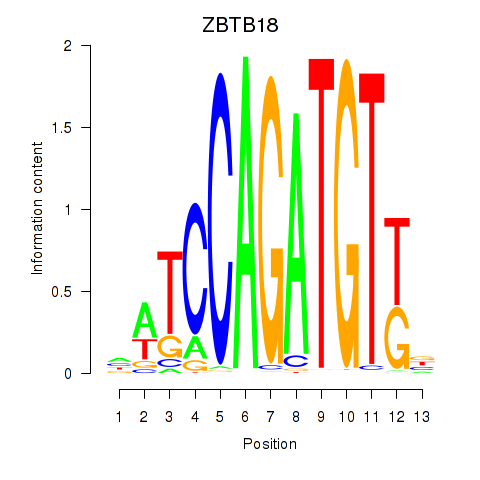
Results for ZBTB18
Z-value: 2.52
Transcription factors associated with ZBTB18
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB18
|
ENSG00000179456.10 | ZBTB18 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB18 | hg38_v1_chr1_+_244051275_244051291 | -0.39 | 2.2e-09 | Click! |
Activity profile of ZBTB18 motif
Sorted Z-values of ZBTB18 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB18
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_188974364 | 26.49 |
ENST00000304636.9
ENST00000317840.9 |
COL3A1
|
collagen type III alpha 1 chain |
| chr17_+_49575828 | 12.69 |
ENST00000328741.6
|
NXPH3
|
neurexophilin 3 |
| chr15_-_89815332 | 12.39 |
ENST00000559874.2
|
ANPEP
|
alanyl aminopeptidase, membrane |
| chr15_-_89814845 | 11.78 |
ENST00000679248.1
ENST00000300060.7 ENST00000560137.2 |
ANPEP
|
alanyl aminopeptidase, membrane |
| chr3_+_100492548 | 10.81 |
ENST00000323523.8
ENST00000403410.5 ENST00000449609.1 |
TMEM45A
|
transmembrane protein 45A |
| chr2_-_162242998 | 10.44 |
ENST00000627638.2
ENST00000447386.5 |
FAP
|
fibroblast activation protein alpha |
| chr2_-_162243375 | 10.36 |
ENST00000188790.9
ENST00000443424.5 |
FAP
|
fibroblast activation protein alpha |
| chr7_+_101127095 | 9.92 |
ENST00000223095.5
|
SERPINE1
|
serpin family E member 1 |
| chr19_+_11538844 | 9.08 |
ENST00000252456.7
|
CNN1
|
calponin 1 |
| chr2_-_1744442 | 9.06 |
ENST00000433670.5
ENST00000425171.1 ENST00000252804.9 |
PXDN
|
peroxidasin |
| chr14_+_94110728 | 8.80 |
ENST00000616764.4
ENST00000618863.1 ENST00000611954.4 ENST00000618200.4 ENST00000621160.4 ENST00000555819.5 ENST00000620396.4 ENST00000612813.4 ENST00000620066.1 |
IFI27
|
interferon alpha inducible protein 27 |
| chr1_-_16980607 | 8.73 |
ENST00000375535.4
|
MFAP2
|
microfibril associated protein 2 |
| chr17_+_79024142 | 8.34 |
ENST00000579760.6
ENST00000339142.6 |
C1QTNF1
|
C1q and TNF related 1 |
| chr17_+_79024243 | 8.10 |
ENST00000311661.4
|
C1QTNF1
|
C1q and TNF related 1 |
| chr19_+_11538767 | 8.06 |
ENST00000592923.5
ENST00000535659.6 |
CNN1
|
calponin 1 |
| chr9_+_122375286 | 8.05 |
ENST00000373698.7
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr1_-_16018005 | 8.03 |
ENST00000406363.2
ENST00000411503.5 ENST00000311890.14 ENST00000487046.1 |
HSPB7
|
heat shock protein family B (small) member 7 |
| chr17_+_34255274 | 7.73 |
ENST00000580907.5
ENST00000225831.4 |
CCL2
|
C-C motif chemokine ligand 2 |
| chr11_+_76783349 | 7.52 |
ENST00000333090.5
|
TSKU
|
tsukushi, small leucine rich proteoglycan |
| chr8_-_48921419 | 7.18 |
ENST00000020945.4
|
SNAI2
|
snail family transcriptional repressor 2 |
| chrX_+_43656289 | 6.15 |
ENST00000338702.4
|
MAOA
|
monoamine oxidase A |
| chr11_+_32091065 | 6.13 |
ENST00000054950.4
|
RCN1
|
reticulocalbin 1 |
| chr17_+_62627628 | 5.92 |
ENST00000303375.10
|
MRC2
|
mannose receptor C type 2 |
| chr1_-_168729187 | 5.70 |
ENST00000367817.4
|
DPT
|
dermatopontin |
| chr11_-_94493798 | 5.61 |
ENST00000538923.1
ENST00000540013.5 ENST00000407439.7 ENST00000323929.8 ENST00000393241.8 |
MRE11
|
MRE11 homolog, double strand break repair nuclease |
| chr17_-_15265230 | 5.59 |
ENST00000676161.1
ENST00000646419.2 ENST00000312280.9 ENST00000494511.7 ENST00000580584.3 ENST00000676221.1 |
PMP22
|
peripheral myelin protein 22 |
| chr9_+_94726657 | 5.51 |
ENST00000375315.8
ENST00000277198.6 ENST00000297979.9 |
AOPEP
|
aminopeptidase O (putative) |
| chr6_-_162727748 | 5.25 |
ENST00000366892.5
ENST00000366898.6 ENST00000366897.5 ENST00000366896.5 ENST00000674250.1 |
PRKN
|
parkin RBR E3 ubiquitin protein ligase |
| chr2_+_33134579 | 5.01 |
ENST00000418533.6
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr2_+_33134620 | 5.00 |
ENST00000402934.5
ENST00000404525.5 ENST00000407925.5 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr2_+_151357583 | 4.66 |
ENST00000243347.5
|
TNFAIP6
|
TNF alpha induced protein 6 |
| chr2_-_88992903 | 4.57 |
ENST00000495489.1
|
IGKV1-8
|
immunoglobulin kappa variable 1-8 |
| chr3_-_160449752 | 4.39 |
ENST00000496222.1
ENST00000471396.1 ENST00000471155.5 ENST00000309784.9 |
TRIM59
|
tripartite motif containing 59 |
| chr17_+_34270213 | 4.24 |
ENST00000378569.2
ENST00000394627.5 ENST00000394630.3 |
CCL7
|
C-C motif chemokine ligand 7 |
| chr12_-_104050112 | 4.22 |
ENST00000547583.1
ENST00000546851.1 ENST00000360814.9 |
GLT8D2
|
glycosyltransferase 8 domain containing 2 |
| chr17_+_50561010 | 4.02 |
ENST00000360761.8
ENST00000354983.8 ENST00000352832.9 |
CACNA1G
|
calcium voltage-gated channel subunit alpha1 G |
| chrX_+_48383516 | 4.00 |
ENST00000620320.4
ENST00000595689.3 |
SSX4
|
SSX family member 4 |
| chr3_+_30606574 | 3.82 |
ENST00000295754.10
ENST00000359013.4 |
TGFBR2
|
transforming growth factor beta receptor 2 |
| chr17_-_69141878 | 3.75 |
ENST00000590645.1
ENST00000284425.7 |
ABCA6
|
ATP binding cassette subfamily A member 6 |
| chr17_+_50560703 | 3.73 |
ENST00000359106.10
|
CACNA1G
|
calcium voltage-gated channel subunit alpha1 G |
| chr4_+_87650277 | 3.73 |
ENST00000339673.11
ENST00000282479.8 |
DMP1
|
dentin matrix acidic phosphoprotein 1 |
| chr16_+_28903359 | 3.68 |
ENST00000564112.1
|
ATP2A1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chrX_-_155334580 | 3.67 |
ENST00000369449.7
ENST00000321926.4 |
CLIC2
|
chloride intracellular channel 2 |
| chr19_-_15808126 | 3.59 |
ENST00000334920.3
|
OR10H1
|
olfactory receptor family 10 subfamily H member 1 |
| chr12_-_47725483 | 3.59 |
ENST00000422538.8
|
ENDOU
|
endonuclease, poly(U) specific |
| chr6_+_162727941 | 3.41 |
ENST00000366888.6
|
PACRG
|
parkin coregulated |
| chr12_-_47725558 | 3.39 |
ENST00000229003.7
|
ENDOU
|
endonuclease, poly(U) specific |
| chr1_-_153545793 | 3.39 |
ENST00000354332.8
ENST00000368716.9 |
S100A4
|
S100 calcium binding protein A4 |
| chr17_-_43022350 | 3.36 |
ENST00000587173.5
ENST00000355653.8 |
VAT1
|
vesicle amine transport 1 |
| chr15_+_92393841 | 3.36 |
ENST00000268164.8
ENST00000539113.5 ENST00000555434.1 |
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr12_-_55728640 | 3.29 |
ENST00000551173.5
ENST00000420846.7 |
CD63
|
CD63 molecule |
| chr12_+_2052977 | 3.27 |
ENST00000399634.6
ENST00000406454.8 ENST00000327702.12 ENST00000347598.9 ENST00000399603.6 ENST00000399641.6 ENST00000399655.6 ENST00000335762.10 ENST00000682835.1 |
CACNA1C
|
calcium voltage-gated channel subunit alpha1 C |
| chr19_-_10231293 | 3.26 |
ENST00000646641.1
|
S1PR2
|
sphingosine-1-phosphate receptor 2 |
| chr6_+_160121859 | 3.24 |
ENST00000324965.8
ENST00000457470.6 |
SLC22A1
|
solute carrier family 22 member 1 |
| chr1_+_8945858 | 3.23 |
ENST00000549778.5
ENST00000377443.7 ENST00000480186.7 ENST00000377436.6 ENST00000377442.3 |
CA6
|
carbonic anhydrase 6 |
| chr6_-_30556477 | 3.20 |
ENST00000376621.8
|
GNL1
|
G protein nucleolar 1 (putative) |
| chr12_-_55727828 | 3.06 |
ENST00000546939.5
|
CD63
|
CD63 molecule |
| chr5_-_151924846 | 3.03 |
ENST00000274576.9
|
GLRA1
|
glycine receptor alpha 1 |
| chr15_+_31326807 | 3.00 |
ENST00000307145.4
|
KLF13
|
Kruppel like factor 13 |
| chr6_+_160121809 | 2.98 |
ENST00000366963.9
|
SLC22A1
|
solute carrier family 22 member 1 |
| chr10_-_93482194 | 2.97 |
ENST00000358334.9
ENST00000371488.3 |
MYOF
|
myoferlin |
| chr17_+_34285734 | 2.91 |
ENST00000305869.4
|
CCL11
|
C-C motif chemokine ligand 11 |
| chr5_-_151924824 | 2.77 |
ENST00000455880.2
|
GLRA1
|
glycine receptor alpha 1 |
| chr17_-_78360066 | 2.77 |
ENST00000587578.1
ENST00000330871.3 |
SOCS3
|
suppressor of cytokine signaling 3 |
| chr11_+_72189659 | 2.70 |
ENST00000393681.6
|
FOLR1
|
folate receptor alpha |
| chr11_-_34357994 | 2.70 |
ENST00000435224.3
|
ABTB2
|
ankyrin repeat and BTB domain containing 2 |
| chrX_+_100644183 | 2.67 |
ENST00000640889.1
ENST00000373004.5 |
SRPX2
|
sushi repeat containing protein X-linked 2 |
| chr12_-_102481744 | 2.54 |
ENST00000644491.1
|
IGF1
|
insulin like growth factor 1 |
| chr4_-_110641920 | 2.47 |
ENST00000354925.6
ENST00000511990.1 ENST00000613094.4 ENST00000614423.4 ENST00000616641.4 ENST00000511837.5 |
PITX2
|
paired like homeodomain 2 |
| chr14_+_64504574 | 2.40 |
ENST00000358738.3
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr10_+_5196831 | 2.39 |
ENST00000263126.3
|
AKR1C4
|
aldo-keto reductase family 1 member C4 |
| chr13_-_33205997 | 2.30 |
ENST00000399365.7
|
STARD13
|
StAR related lipid transfer domain containing 13 |
| chr19_+_40613416 | 2.29 |
ENST00000599724.5
ENST00000597071.5 |
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr11_-_119381629 | 2.27 |
ENST00000260187.7
ENST00000455332.6 |
USP2
|
ubiquitin specific peptidase 2 |
| chr9_+_113501359 | 2.23 |
ENST00000343817.9
ENST00000394646.7 |
RGS3
|
regulator of G protein signaling 3 |
| chr5_+_132673983 | 2.23 |
ENST00000622422.1
ENST00000231449.7 ENST00000350025.2 |
IL4
|
interleukin 4 |
| chr12_-_55728339 | 2.18 |
ENST00000552754.5
|
CD63
|
CD63 molecule |
| chr6_+_41637005 | 2.08 |
ENST00000419164.6
ENST00000373051.6 |
MDFI
|
MyoD family inhibitor |
| chr19_-_51645545 | 2.07 |
ENST00000534261.3
|
SIGLEC5
|
sialic acid binding Ig like lectin 5 |
| chr2_-_98731063 | 2.04 |
ENST00000393487.6
|
MGAT4A
|
alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase A |
| chr2_+_113127588 | 2.03 |
ENST00000409930.4
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr6_-_165663170 | 1.99 |
ENST00000539869.4
|
PDE10A
|
phosphodiesterase 10A |
| chr12_-_55727796 | 1.97 |
ENST00000550776.5
|
CD63
|
CD63 molecule |
| chrX_-_48196763 | 1.96 |
ENST00000311798.5
ENST00000347757.6 |
SSX5
|
SSX family member 5 |
| chr3_+_124584625 | 1.96 |
ENST00000291478.9
ENST00000682363.1 ENST00000454902.1 |
KALRN
|
kalirin RhoGEF kinase |
| chr14_-_77271200 | 1.93 |
ENST00000298352.5
|
NGB
|
neuroglobin |
| chr12_+_2053311 | 1.86 |
ENST00000399617.6
ENST00000683482.1 |
CACNA1C
|
calcium voltage-gated channel subunit alpha1 C |
| chr10_-_69573416 | 1.80 |
ENST00000242462.5
|
NEUROG3
|
neurogenin 3 |
| chr5_-_149944744 | 1.76 |
ENST00000255266.10
ENST00000617647.4 ENST00000613228.1 |
PDE6A
|
phosphodiesterase 6A |
| chr16_-_29862890 | 1.75 |
ENST00000563415.1
|
CDIPT
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| chr20_+_59300547 | 1.75 |
ENST00000644821.1
|
EDN3
|
endothelin 3 |
| chr1_-_156677400 | 1.71 |
ENST00000368223.4
|
NES
|
nestin |
| chr3_+_121593363 | 1.71 |
ENST00000338040.6
|
FBXO40
|
F-box protein 40 |
| chr21_-_34511243 | 1.69 |
ENST00000399284.1
|
KCNE1
|
potassium voltage-gated channel subfamily E regulatory subunit 1 |
| chr17_-_63972918 | 1.69 |
ENST00000435607.3
|
SCN4A
|
sodium voltage-gated channel alpha subunit 4 |
| chr7_-_95324524 | 1.68 |
ENST00000222381.8
|
PON1
|
paraoxonase 1 |
| chr1_+_186296267 | 1.67 |
ENST00000533951.5
ENST00000367482.8 ENST00000635041.1 ENST00000367483.8 ENST00000367485.4 ENST00000445192.7 |
PRG4
|
proteoglycan 4 |
| chr20_+_59300703 | 1.64 |
ENST00000395654.3
|
EDN3
|
endothelin 3 |
| chr4_+_155903688 | 1.62 |
ENST00000536354.3
|
TDO2
|
tryptophan 2,3-dioxygenase |
| chr2_-_89117844 | 1.53 |
ENST00000490686.1
|
IGKV1-17
|
immunoglobulin kappa variable 1-17 |
| chrX_-_111270474 | 1.51 |
ENST00000324068.2
|
CAPN6
|
calpain 6 |
| chr1_-_33182030 | 1.51 |
ENST00000291416.10
|
TRIM62
|
tripartite motif containing 62 |
| chr19_+_50432885 | 1.43 |
ENST00000357701.6
|
MYBPC2
|
myosin binding protein C2 |
| chr4_+_74445126 | 1.42 |
ENST00000395748.8
|
AREG
|
amphiregulin |
| chr20_+_59300589 | 1.42 |
ENST00000337938.7
ENST00000371025.7 |
EDN3
|
endothelin 3 |
| chr21_-_34511317 | 1.40 |
ENST00000611936.1
ENST00000337385.7 |
KCNE1
|
potassium voltage-gated channel subfamily E regulatory subunit 1 |
| chr11_+_60056653 | 1.39 |
ENST00000278865.8
|
MS4A3
|
membrane spanning 4-domains A3 |
| chr10_+_96304392 | 1.35 |
ENST00000630152.1
|
DNTT
|
DNA nucleotidylexotransferase |
| chr17_-_19748341 | 1.34 |
ENST00000395555.7
|
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1 |
| chr11_+_60056587 | 1.33 |
ENST00000395032.6
ENST00000358152.6 |
MS4A3
|
membrane spanning 4-domains A3 |
| chr15_-_74209019 | 1.33 |
ENST00000323940.9
|
STRA6
|
signaling receptor and transporter of retinol STRA6 |
| chr4_+_74445302 | 1.29 |
ENST00000502307.1
|
AREG
|
amphiregulin |
| chr2_-_88966767 | 1.25 |
ENST00000464162.1
|
IGKV1-6
|
immunoglobulin kappa variable 1-6 |
| chr12_+_112125531 | 1.24 |
ENST00000549358.5
ENST00000257604.9 ENST00000548092.5 ENST00000412615.7 ENST00000552896.1 |
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chr17_-_19748285 | 1.12 |
ENST00000570414.1
ENST00000225740.11 |
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1 |
| chr15_-_74208969 | 1.12 |
ENST00000423167.6
ENST00000432245.6 |
STRA6
|
signaling receptor and transporter of retinol STRA6 |
| chr10_+_96304425 | 1.10 |
ENST00000371174.5
|
DNTT
|
DNA nucleotidylexotransferase |
| chr20_+_59300402 | 1.07 |
ENST00000311585.11
ENST00000371028.6 |
EDN3
|
endothelin 3 |
| chr15_-_31161157 | 1.04 |
ENST00000542188.5
|
TRPM1
|
transient receptor potential cation channel subfamily M member 1 |
| chr4_+_47485268 | 1.01 |
ENST00000273859.8
ENST00000504445.1 |
ATP10D
|
ATPase phospholipid transporting 10D (putative) |
| chr7_+_121328991 | 0.99 |
ENST00000222462.3
|
WNT16
|
Wnt family member 16 |
| chr1_+_26410809 | 0.97 |
ENST00000254231.4
ENST00000326279.11 |
LIN28A
|
lin-28 homolog A |
| chr11_-_94493863 | 0.96 |
ENST00000323977.7
ENST00000536754.5 |
MRE11
|
MRE11 homolog, double strand break repair nuclease |
| chr1_-_207032749 | 0.96 |
ENST00000359470.6
ENST00000461135.2 |
C1orf116
|
chromosome 1 open reading frame 116 |
| chr12_-_53727476 | 0.94 |
ENST00000549784.5
ENST00000262059.8 |
CALCOCO1
|
calcium binding and coiled-coil domain 1 |
| chr14_+_67241278 | 0.93 |
ENST00000676464.1
|
MPP5
|
membrane palmitoylated protein 5 |
| chr2_-_74465162 | 0.91 |
ENST00000649854.1
ENST00000650523.1 ENST00000649601.1 ENST00000448666.7 ENST00000409065.5 ENST00000414701.1 ENST00000452063.7 ENST00000649075.1 ENST00000648810.1 ENST00000462443.2 |
MOGS
|
mannosyl-oligosaccharide glucosidase |
| chr18_+_54828406 | 0.88 |
ENST00000262094.10
|
RAB27B
|
RAB27B, member RAS oncogene family |
| chr7_+_95485934 | 0.86 |
ENST00000325885.6
|
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr13_+_113122791 | 0.84 |
ENST00000375559.8
ENST00000409306.5 ENST00000375551.7 |
F10
|
coagulation factor X |
| chrX_-_102516714 | 0.82 |
ENST00000289373.5
|
TMSB15A
|
thymosin beta 15A |
| chrX_+_150361559 | 0.80 |
ENST00000262858.8
|
MAMLD1
|
mastermind like domain containing 1 |
| chr14_+_67240713 | 0.80 |
ENST00000677382.1
|
MPP5
|
membrane palmitoylated protein 5 |
| chr1_-_92486049 | 0.78 |
ENST00000427103.5
|
GFI1
|
growth factor independent 1 transcriptional repressor |
| chr11_-_62556230 | 0.70 |
ENST00000530285.5
|
AHNAK
|
AHNAK nucleoprotein |
| chr1_+_37556913 | 0.67 |
ENST00000296218.8
ENST00000652629.1 |
DNALI1
|
dynein axonemal light intermediate chain 1 |
| chr1_+_204073104 | 0.67 |
ENST00000367204.6
|
SOX13
|
SRY-box transcription factor 13 |
| chr11_-_101583985 | 0.62 |
ENST00000344327.8
|
TRPC6
|
transient receptor potential cation channel subfamily C member 6 |
| chr21_-_32813695 | 0.62 |
ENST00000479548.2
ENST00000490358.5 |
C21orf62
|
chromosome 21 open reading frame 62 |
| chr11_+_73289403 | 0.61 |
ENST00000535931.2
ENST00000544437.6 |
P2RY6
|
pyrimidinergic receptor P2Y6 |
| chr10_-_22003678 | 0.60 |
ENST00000376980.8
|
DNAJC1
|
DnaJ heat shock protein family (Hsp40) member C1 |
| chr17_+_9021501 | 0.58 |
ENST00000173229.7
|
NTN1
|
netrin 1 |
| chr2_-_157874976 | 0.57 |
ENST00000682025.1
ENST00000683487.1 ENST00000682300.1 ENST00000683441.1 ENST00000684595.1 ENST00000683426.1 ENST00000683820.1 ENST00000263640.7 |
ACVR1
|
activin A receptor type 1 |
| chr18_+_79863668 | 0.57 |
ENST00000316249.3
|
KCNG2
|
potassium voltage-gated channel modifier subfamily G member 2 |
| chr20_-_63361337 | 0.56 |
ENST00000370263.9
|
CHRNA4
|
cholinergic receptor nicotinic alpha 4 subunit |
| chr17_-_19748355 | 0.56 |
ENST00000494157.6
|
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1 |
| chr1_+_23743462 | 0.56 |
ENST00000609199.1
|
ELOA
|
elongin A |
| chr12_-_53499615 | 0.53 |
ENST00000267079.6
|
MAP3K12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr14_-_91244669 | 0.50 |
ENST00000650645.1
|
GPR68
|
G protein-coupled receptor 68 |
| chr8_+_24384275 | 0.49 |
ENST00000256412.8
|
ADAMDEC1
|
ADAM like decysin 1 |
| chr7_-_157010615 | 0.43 |
ENST00000252971.11
|
MNX1
|
motor neuron and pancreas homeobox 1 |
| chr8_+_24384455 | 0.38 |
ENST00000522298.1
|
ADAMDEC1
|
ADAM like decysin 1 |
| chr12_+_53985783 | 0.38 |
ENST00000513209.1
|
ENSG00000273049.1
|
novel protein, readthrough between HOXC10 and HOXC5 |
| chr3_-_111595339 | 0.38 |
ENST00000317012.5
|
ZBED2
|
zinc finger BED-type containing 2 |
| chr9_-_13165442 | 0.36 |
ENST00000542239.1
ENST00000538841.5 ENST00000433359.6 |
MPDZ
|
multiple PDZ domain crumbs cell polarity complex component |
| chr12_+_1629197 | 0.29 |
ENST00000397196.7
|
WNT5B
|
Wnt family member 5B |
| chr17_+_44349050 | 0.27 |
ENST00000639447.1
|
GRN
|
granulin precursor |
| chr14_-_64504570 | 0.22 |
ENST00000394715.1
|
ZBTB25
|
zinc finger and BTB domain containing 25 |
| chr10_+_103493931 | 0.21 |
ENST00000369780.8
|
NEURL1
|
neuralized E3 ubiquitin protein ligase 1 |
| chr12_+_25973748 | 0.19 |
ENST00000542865.5
|
RASSF8
|
Ras association domain family member 8 |
| chr21_-_32813679 | 0.17 |
ENST00000487113.1
ENST00000382373.4 |
C21orf62
|
chromosome 21 open reading frame 62 |
| chr7_+_95485898 | 0.15 |
ENST00000428113.5
|
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr13_-_60013178 | 0.10 |
ENST00000498416.2
ENST00000465066.5 |
DIAPH3
|
diaphanous related formin 3 |
| chr10_+_70052582 | 0.09 |
ENST00000676699.1
|
MACROH2A2
|
macroH2A.2 histone |
| chr7_+_90596281 | 0.03 |
ENST00000380050.8
|
CDK14
|
cyclin dependent kinase 14 |
| chr2_-_65432591 | 0.03 |
ENST00000356388.9
|
SPRED2
|
sprouty related EVH1 domain containing 2 |
| chr7_+_97732046 | 0.01 |
ENST00000350485.8
ENST00000346867.4 ENST00000319273.10 |
TAC1
|
tachykinin precursor 1 |
| chr12_-_53499444 | 0.01 |
ENST00000547488.6
|
MAP3K12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr11_+_63888515 | 0.01 |
ENST00000509502.6
ENST00000512060.1 |
MARK2
|
microtubule affinity regulating kinase 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 20.8 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 4.4 | 26.5 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 3.3 | 9.9 | GO:2000097 | chronological cell aging(GO:0001300) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 3.3 | 16.4 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 2.6 | 7.7 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 2.4 | 7.2 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) negative regulation of response to alcohol(GO:1901420) |
| 1.9 | 7.5 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 1.7 | 5.2 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 1.5 | 8.7 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 1.4 | 10.0 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 1.3 | 3.8 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 1.2 | 3.7 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.9 | 6.6 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.9 | 17.1 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.9 | 2.7 | GO:0060974 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) |
| 0.9 | 6.2 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.8 | 5.9 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.8 | 5.8 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.8 | 2.5 | GO:0060577 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.8 | 12.9 | GO:0086045 | membrane depolarization during AV node cell action potential(GO:0086045) |
| 0.8 | 10.5 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.7 | 2.2 | GO:0070343 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) cellular response to mercury ion(GO:0071288) |
| 0.7 | 2.9 | GO:0035962 | response to interleukin-13(GO:0035962) |
| 0.7 | 4.2 | GO:2000501 | regulation of natural killer cell chemotaxis(GO:2000501) positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.7 | 2.8 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.6 | 29.7 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.5 | 2.5 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.5 | 3.4 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.5 | 2.3 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.4 | 1.7 | GO:0035750 | protein localization to myelin sheath abaxonal region(GO:0035750) |
| 0.4 | 8.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.4 | 1.7 | GO:0034443 | negative regulation of lipoprotein oxidation(GO:0034443) regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) response to fluoride(GO:1902617) |
| 0.4 | 3.7 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.4 | 2.4 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.4 | 2.4 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.4 | 2.7 | GO:0060750 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.4 | 6.1 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.4 | 3.3 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.3 | 3.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.3 | 1.0 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) |
| 0.3 | 1.0 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.3 | 5.6 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.3 | 9.1 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.3 | 1.6 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.3 | 0.8 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.3 | 2.0 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.2 | 3.0 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.2 | 3.7 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.2 | 2.7 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 2.0 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.2 | 3.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.2 | 3.1 | GO:1902260 | negative regulation of protein targeting to membrane(GO:0090315) negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.2 | 0.6 | GO:0061445 | endocardial cell fate commitment(GO:0060957) endocardial cushion cell differentiation(GO:0061443) endocardial cushion cell fate commitment(GO:0061445) |
| 0.2 | 8.8 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.2 | 4.7 | GO:0030728 | ovulation(GO:0030728) |
| 0.2 | 4.4 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 2.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 2.1 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.1 | 12.7 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 5.7 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 3.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 1.8 | GO:0060290 | transdifferentiation(GO:0060290) |
| 0.1 | 1.5 | GO:0046596 | regulation of viral entry into host cell(GO:0046596) |
| 0.1 | 1.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.5 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 1.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.6 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.7 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.1 | 2.3 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.1 | 0.8 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 2.7 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.1 | 2.0 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 2.0 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 7.0 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 5.9 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 0.5 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 1.0 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.1 | 1.7 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) |
| 0.1 | 2.4 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 2.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 0.6 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 11.4 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 0.6 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 4.2 | GO:0000271 | polysaccharide biosynthetic process(GO:0000271) |
| 0.0 | 0.4 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 1.0 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.6 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 1.0 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 3.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 1.7 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 3.0 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 0.2 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 1.8 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.3 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 3.4 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 1.2 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.0 | 1.5 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.7 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.9 | GO:0009311 | oligosaccharide metabolic process(GO:0009311) |
| 0.0 | 1.5 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 3.8 | GO:0006869 | lipid transport(GO:0006869) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 26.5 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 1.4 | 20.8 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.9 | 18.7 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.9 | 10.5 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.9 | 5.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.6 | 3.8 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.6 | 6.6 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.4 | 5.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.3 | 2.5 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.3 | 1.0 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.3 | 5.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 3.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.2 | 7.7 | GO:0043218 | compact myelin(GO:0043218) |
| 0.2 | 0.9 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.2 | 16.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 4.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.2 | 29.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.2 | 8.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 3.7 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 0.6 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 1.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 8.8 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 1.7 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.1 | 9.9 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 7.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 3.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 4.7 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 1.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 22.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 0.6 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 2.4 | GO:0000791 | euchromatin(GO:0000791) |
| 0.1 | 2.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 1.4 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.6 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 3.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 3.6 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 3.0 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 55.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 9.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 23.8 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 2.7 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 4.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 3.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 2.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.1 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 6.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 6.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 3.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 1.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.7 | GO:0030286 | dynein complex(GO:0030286) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.8 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 2.6 | 7.7 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 2.2 | 11.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 2.0 | 26.5 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 2.0 | 10.0 | GO:0050436 | microfibril binding(GO:0050436) |
| 1.6 | 8.0 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 1.6 | 6.2 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 1.3 | 3.8 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 1.2 | 29.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 1.2 | 5.8 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 1.0 | 5.9 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.8 | 4.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.8 | 20.8 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.7 | 2.2 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.7 | 2.9 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.7 | 2.7 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.6 | 6.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.6 | 2.4 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.6 | 8.7 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.5 | 1.6 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.5 | 3.0 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.5 | 3.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.5 | 5.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.5 | 5.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.4 | 1.7 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.4 | 6.6 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.3 | 8.0 | GO:0031005 | filamin binding(GO:0031005) |
| 0.3 | 8.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.3 | 2.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.3 | 3.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.2 | 3.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.2 | 22.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 3.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 4.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 2.0 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.2 | 3.6 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.2 | 3.4 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.2 | 2.9 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.1 | 1.9 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 3.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 3.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 2.1 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.6 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.1 | 1.7 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 9.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 3.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 3.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 1.8 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 2.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 1.0 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 2.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.9 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.1 | 2.5 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 17.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 2.4 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.1 | 1.7 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 0.7 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 2.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 2.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.9 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 7.2 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 4.8 | GO:0043492 | ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 1.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 1.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 7.3 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 2.3 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 2.8 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.6 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 1.8 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 8.0 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 11.4 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 1.7 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 2.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 3.0 | GO:0016757 | transferase activity, transferring glycosyl groups(GO:0016757) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 26.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 55.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 9.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 10.5 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.2 | 24.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 3.8 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 5.6 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 2.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 2.2 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 17.4 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 11.3 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 6.5 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 3.3 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.1 | 2.9 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 16.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 2.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 2.0 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 2.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 5.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 1.8 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 3.0 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.5 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 9.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.7 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 37.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.3 | 6.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.3 | 6.1 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.2 | 1.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 14.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 3.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 2.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.2 | 9.9 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.2 | 3.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 3.8 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 2.8 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 4.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 5.8 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 8.0 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 0.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 2.0 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 2.0 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 8.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 5.9 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 3.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 0.6 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 10.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 0.9 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 1.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.8 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 5.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 3.6 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 1.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.8 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.6 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 2.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 3.8 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.6 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |