Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
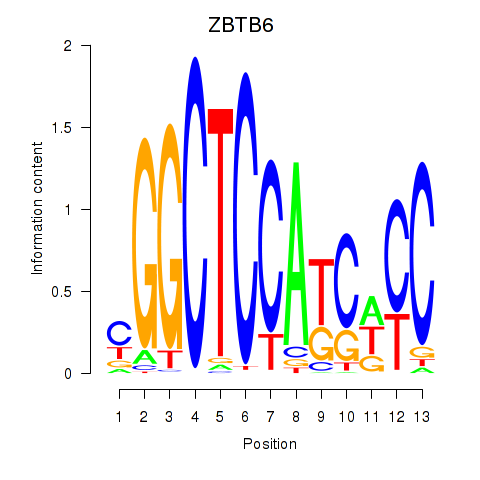
Results for ZBTB6
Z-value: 7.92
Transcription factors associated with ZBTB6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB6
|
ENSG00000186130.5 | ZBTB6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB6 | hg38_v1_chr9_-_122913299_122913333 | 0.14 | 4.5e-02 | Click! |
Activity profile of ZBTB6 motif
Sorted Z-values of ZBTB6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.9 | 53.1 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 6.3 | 6.3 | GO:0032058 | positive regulation of translation in response to stress(GO:0032056) positive regulation of translational initiation in response to stress(GO:0032058) |
| 6.3 | 18.9 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 5.3 | 21.2 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) negative regulation of type B pancreatic cell development(GO:2000077) |
| 5.2 | 20.9 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 5.0 | 19.8 | GO:1904980 | positive regulation of endosome organization(GO:1904980) |
| 4.5 | 40.7 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 3.8 | 30.5 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 3.6 | 18.2 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 3.6 | 10.8 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 3.2 | 9.7 | GO:0036048 | protein demalonylation(GO:0036046) peptidyl-lysine demalonylation(GO:0036047) protein desuccinylation(GO:0036048) peptidyl-lysine desuccinylation(GO:0036049) protein deglutarylation(GO:0061698) peptidyl-lysine deglutarylation(GO:0061699) |
| 3.2 | 9.6 | GO:0051939 | gamma-aminobutyric acid import(GO:0051939) |
| 3.2 | 9.6 | GO:1904692 | positive regulation of type B pancreatic cell proliferation(GO:1904692) |
| 2.7 | 10.9 | GO:0019086 | late viral transcription(GO:0019086) |
| 2.7 | 8.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 2.6 | 10.3 | GO:0048627 | myoblast development(GO:0048627) |
| 2.5 | 7.6 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 2.5 | 9.8 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 2.4 | 7.3 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 2.2 | 15.4 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 2.1 | 8.6 | GO:1904450 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 2.1 | 6.4 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 2.1 | 8.4 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 2.1 | 12.6 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 2.1 | 6.2 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 1.9 | 7.7 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 1.9 | 7.5 | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) |
| 1.9 | 18.6 | GO:1905146 | lysosomal protein catabolic process(GO:1905146) |
| 1.9 | 5.6 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 1.9 | 7.4 | GO:1903971 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 1.9 | 11.2 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 1.8 | 5.5 | GO:0071284 | establishment of blood-nerve barrier(GO:0008065) cellular response to lead ion(GO:0071284) positive regulation of bicellular tight junction assembly(GO:1903348) |
| 1.8 | 9.1 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 1.8 | 8.8 | GO:0030070 | insulin processing(GO:0030070) |
| 1.8 | 8.8 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 1.7 | 5.2 | GO:0071529 | cementum mineralization(GO:0071529) |
| 1.7 | 6.7 | GO:0032811 | negative regulation of epinephrine secretion(GO:0032811) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 1.6 | 4.9 | GO:0036146 | cellular response to mycotoxin(GO:0036146) |
| 1.6 | 8.1 | GO:2001074 | regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 1.6 | 11.3 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 1.6 | 8.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 1.6 | 11.1 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 1.6 | 7.9 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 1.4 | 4.3 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 1.4 | 12.8 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 1.4 | 5.5 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 1.4 | 4.1 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 1.4 | 4.1 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 1.3 | 8.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 1.3 | 3.9 | GO:0033602 | negative regulation of dopamine secretion(GO:0033602) |
| 1.3 | 6.4 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 1.3 | 3.8 | GO:1903465 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 1.2 | 2.5 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 1.2 | 3.7 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 1.2 | 3.6 | GO:0018282 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 1.2 | 20.4 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 1.2 | 7.0 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 1.2 | 16.3 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 1.1 | 2.3 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 1.1 | 8.0 | GO:0035093 | piRNA metabolic process(GO:0034587) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 1.1 | 8.8 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 1.1 | 3.3 | GO:0014876 | negative regulation of muscle atrophy(GO:0014736) response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 1.1 | 6.4 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 1.1 | 3.2 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 1.0 | 4.0 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 1.0 | 6.8 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 1.0 | 2.9 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.9 | 3.8 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.9 | 3.8 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.9 | 2.8 | GO:0035048 | splicing factor protein import into nucleus(GO:0035048) |
| 0.9 | 8.3 | GO:0090487 | toxin catabolic process(GO:0009407) secondary metabolite catabolic process(GO:0090487) |
| 0.9 | 4.5 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.9 | 15.3 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.9 | 13.4 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.9 | 51.3 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.9 | 2.6 | GO:0060128 | corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.9 | 3.4 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.9 | 6.0 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.8 | 1.7 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.8 | 2.5 | GO:1900155 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.8 | 5.0 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.8 | 5.8 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.8 | 12.3 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.8 | 10.6 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.8 | 18.5 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.8 | 4.0 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.8 | 8.6 | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.8 | 12.9 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.8 | 15.1 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.8 | 21.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.8 | 2.3 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.7 | 1.5 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.7 | 11.0 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.7 | 5.8 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.7 | 7.8 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.7 | 2.8 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.7 | 2.1 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.7 | 11.9 | GO:2000050 | regulation of non-canonical Wnt signaling pathway(GO:2000050) |
| 0.7 | 12.6 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.7 | 9.0 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.7 | 2.1 | GO:0048382 | mesendoderm development(GO:0048382) |
| 0.7 | 4.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) activation of phospholipase A2 activity(GO:0032431) |
| 0.7 | 6.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.7 | 13.5 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.7 | 2.0 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.7 | 7.3 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.7 | 2.0 | GO:0072244 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.7 | 7.8 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.6 | 3.9 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.6 | 2.6 | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.6 | 17.3 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.6 | 1.9 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.6 | 3.8 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.6 | 3.8 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.6 | 1.9 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.6 | 3.2 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.6 | 6.2 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.6 | 9.9 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.6 | 6.8 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.6 | 7.4 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.6 | 1.2 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.6 | 3.0 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.6 | 7.3 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.6 | 9.0 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.6 | 3.0 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.6 | 8.9 | GO:0021794 | thalamus development(GO:0021794) |
| 0.6 | 6.9 | GO:0009629 | response to gravity(GO:0009629) |
| 0.6 | 3.5 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.6 | 9.8 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.6 | 2.9 | GO:0071503 | response to heparin(GO:0071503) cellular response to heparin(GO:0071504) |
| 0.6 | 4.0 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.6 | 2.3 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.6 | 17.5 | GO:0001825 | blastocyst formation(GO:0001825) |
| 0.6 | 17.4 | GO:0021772 | olfactory bulb development(GO:0021772) |
| 0.6 | 3.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.5 | 4.9 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.5 | 6.8 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.5 | 30.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.5 | 5.1 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.5 | 7.6 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.5 | 4.4 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.5 | 5.9 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.5 | 2.4 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.5 | 6.6 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.5 | 2.4 | GO:0089712 | L-aspartate transport(GO:0070778) L-aspartate transmembrane transport(GO:0089712) |
| 0.5 | 4.2 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.5 | 3.7 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.5 | 4.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.5 | 13.8 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.5 | 10.1 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.4 | 1.8 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) response to fluoride(GO:1902617) |
| 0.4 | 3.1 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.4 | 3.1 | GO:0007440 | foregut morphogenesis(GO:0007440) embryonic foregut morphogenesis(GO:0048617) |
| 0.4 | 3.0 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.4 | 2.6 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.4 | 17.9 | GO:0014072 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.4 | 11.5 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.4 | 6.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.4 | 2.1 | GO:1901906 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.4 | 1.2 | GO:0048880 | sensory system development(GO:0048880) |
| 0.4 | 8.0 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.4 | 7.1 | GO:0060022 | hard palate development(GO:0060022) |
| 0.4 | 3.1 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.4 | 1.5 | GO:0018032 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 0.4 | 5.3 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.4 | 1.9 | GO:0033504 | floor plate development(GO:0033504) |
| 0.4 | 6.8 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.4 | 3.4 | GO:0046078 | dUMP metabolic process(GO:0046078) |
| 0.4 | 1.1 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.4 | 5.7 | GO:0008212 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.4 | 3.5 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.3 | 8.0 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.3 | 1.7 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.3 | 1.0 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.3 | 1.7 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.3 | 2.4 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.3 | 3.7 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.3 | 2.3 | GO:0060751 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.3 | 0.6 | GO:0051586 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.3 | 3.2 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.3 | 4.4 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.3 | 0.6 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.3 | 2.4 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.3 | 15.7 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.3 | 3.5 | GO:0050665 | hydrogen peroxide biosynthetic process(GO:0050665) |
| 0.3 | 3.5 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.3 | 0.9 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.3 | 4.3 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.3 | 0.9 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.3 | 4.6 | GO:0048535 | lymph node development(GO:0048535) |
| 0.3 | 1.1 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.3 | 6.1 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.3 | 9.7 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.3 | 4.7 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.3 | 10.0 | GO:0033014 | tetrapyrrole biosynthetic process(GO:0033014) |
| 0.3 | 5.5 | GO:0007614 | short-term memory(GO:0007614) |
| 0.2 | 7.0 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.2 | 2.0 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.2 | 9.3 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.2 | 1.7 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.2 | 8.0 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.2 | 4.0 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.2 | 1.4 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.2 | 1.8 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.2 | 4.4 | GO:0010666 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.2 | 4.1 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.2 | 1.6 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.2 | 4.0 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.2 | 5.5 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.2 | 1.8 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.2 | 7.8 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.2 | 2.5 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.2 | 1.0 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.2 | 7.1 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.2 | 12.2 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.2 | 0.4 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.2 | 0.4 | GO:0060526 | prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.2 | 4.8 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.2 | 3.9 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.2 | 3.5 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.2 | 7.4 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.2 | 4.6 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.2 | 4.3 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.2 | 21.5 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.2 | 12.1 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.2 | 12.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.2 | 5.2 | GO:0009083 | branched-chain amino acid metabolic process(GO:0009081) branched-chain amino acid catabolic process(GO:0009083) |
| 0.2 | 3.0 | GO:0006505 | GPI anchor metabolic process(GO:0006505) GPI anchor biosynthetic process(GO:0006506) |
| 0.2 | 4.4 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.2 | 1.5 | GO:0021815 | cell motility involved in cerebral cortex radial glia guided migration(GO:0021814) modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) |
| 0.2 | 2.9 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.2 | 1.3 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.2 | 1.3 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 5.0 | GO:0014898 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.2 | 0.6 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 2.4 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 6.5 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.1 | 0.7 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.1 | 5.1 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.1 | 5.1 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 3.9 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 5.5 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 3.2 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.1 | 6.6 | GO:0030317 | sperm motility(GO:0030317) |
| 0.1 | 5.2 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.1 | 22.4 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 4.9 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 6.6 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 2.9 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 | 4.0 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.1 | 3.0 | GO:0015800 | acidic amino acid transport(GO:0015800) |
| 0.1 | 5.7 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 1.2 | GO:1904646 | cellular response to beta-amyloid(GO:1904646) |
| 0.1 | 4.1 | GO:0035904 | aorta development(GO:0035904) |
| 0.1 | 1.0 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.1 | 17.7 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 1.3 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 1.1 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 3.2 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.1 | 7.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 1.3 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 3.9 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.1 | 0.9 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 5.7 | GO:0051646 | mitochondrion localization(GO:0051646) |
| 0.1 | 0.4 | GO:0032904 | viral protein processing(GO:0019082) negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) regulation of neurotrophin production(GO:0032899) negative regulation of neurotrophin production(GO:0032900) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.1 | 3.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 1.4 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.1 | 3.9 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 3.1 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 3.3 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.1 | 3.9 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.1 | 13.7 | GO:0030518 | intracellular steroid hormone receptor signaling pathway(GO:0030518) |
| 0.1 | 1.2 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.1 | 1.1 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.1 | 2.5 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.1 | 4.9 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.1 | 4.2 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.1 | 1.8 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 6.9 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 0.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.1 | 4.6 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 4.5 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.1 | 0.9 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 2.4 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.1 | 2.1 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 2.4 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 1.6 | GO:0007099 | centriole replication(GO:0007099) |
| 0.1 | 0.5 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 1.8 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.1 | 3.4 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 1.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 1.1 | GO:0060563 | neuroepithelial cell differentiation(GO:0060563) |
| 0.1 | 2.1 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 6.7 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.0 | 6.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.7 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 2.1 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 2.8 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 1.0 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 2.4 | GO:0008585 | female gonad development(GO:0008585) |
| 0.0 | 0.3 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.8 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 2.3 | GO:0000271 | polysaccharide biosynthetic process(GO:0000271) |
| 0.0 | 1.3 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 1.3 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.0 | 3.5 | GO:0003014 | renal system process(GO:0003014) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.6 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 2.8 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 2.4 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.0 | 0.5 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.1 | GO:0002911 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.0 | 0.1 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.8 | GO:0005977 | glycogen metabolic process(GO:0005977) |
| 0.0 | 1.0 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 4.5 | GO:0048232 | spermatogenesis(GO:0007283) male gamete generation(GO:0048232) |
| 0.0 | 0.2 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.1 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 18.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 2.1 | 8.6 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 2.1 | 6.4 | GO:0034657 | GID complex(GO:0034657) |
| 1.9 | 32.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 1.4 | 5.5 | GO:0019034 | viral replication complex(GO:0019034) |
| 1.3 | 8.0 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 1.3 | 15.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 1.2 | 7.4 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 1.1 | 3.4 | GO:0031251 | PAN complex(GO:0031251) |
| 1.1 | 13.6 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 1.1 | 12.8 | GO:0016342 | catenin complex(GO:0016342) |
| 1.1 | 23.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 1.1 | 8.4 | GO:0034464 | BBSome(GO:0034464) |
| 1.0 | 5.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 1.0 | 5.8 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.9 | 7.6 | GO:0060203 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.9 | 8.0 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.9 | 7.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.9 | 6.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.9 | 8.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.8 | 18.6 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.8 | 4.7 | GO:0002177 | manchette(GO:0002177) |
| 0.8 | 3.8 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.7 | 5.2 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.7 | 7.8 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.7 | 5.6 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.7 | 28.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.7 | 4.1 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.7 | 4.0 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.7 | 13.1 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.7 | 2.0 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.6 | 7.0 | GO:0005638 | lamin filament(GO:0005638) |
| 0.6 | 6.8 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.5 | 7.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.5 | 6.5 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.5 | 3.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.5 | 12.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.5 | 5.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.5 | 16.4 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.5 | 0.5 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.4 | 1.8 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.4 | 7.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.4 | 2.2 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.4 | 3.9 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.4 | 2.1 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.4 | 2.5 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.4 | 4.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.4 | 2.0 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.4 | 4.5 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.4 | 3.7 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.3 | 3.8 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.3 | 5.5 | GO:0000800 | lateral element(GO:0000800) |
| 0.3 | 18.5 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.3 | 7.9 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.3 | 6.5 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.3 | 1.5 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.3 | 7.1 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.3 | 4.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.3 | 10.8 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.3 | 18.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.3 | 43.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.3 | 7.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.3 | 6.9 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.3 | 6.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.3 | 16.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 7.0 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.2 | 1.5 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.2 | 11.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.2 | 11.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 13.2 | GO:0005844 | polysome(GO:0005844) |
| 0.2 | 5.9 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.2 | 16.6 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.2 | 1.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.2 | 6.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.2 | 24.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.2 | 14.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.2 | 3.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 9.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.2 | 1.0 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.2 | 2.8 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.2 | 9.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 16.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.2 | 1.7 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 9.6 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 19.7 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 18.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 5.0 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 1.8 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.1 | 8.2 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 5.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 1.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 1.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 10.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 5.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 6.3 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 8.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 7.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 4.4 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 5.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 9.5 | GO:0009986 | cell surface(GO:0009986) |
| 0.1 | 2.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 13.3 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 1.7 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 1.1 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 100.4 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 3.3 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 2.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.8 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 6.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.7 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 2.0 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 3.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 2.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.8 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 1.6 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 3.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.8 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 1.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 41.8 | GO:0016021 | integral component of membrane(GO:0016021) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.9 | 53.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 4.0 | 11.9 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 3.6 | 18.2 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 3.5 | 10.6 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 3.2 | 9.7 | GO:0061697 | protein-malonyllysine demalonylase activity(GO:0036054) protein-succinyllysine desuccinylase activity(GO:0036055) protein-glutaryllysine deglutarylase activity(GO:0061697) |
| 3.0 | 12.2 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 2.8 | 19.8 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 2.7 | 8.0 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 2.6 | 12.9 | GO:0050610 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 2.6 | 7.7 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 2.5 | 10.2 | GO:0052812 | phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 2.5 | 15.2 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 2.2 | 6.7 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 2.1 | 8.6 | GO:0047298 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 2.1 | 55.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 2.0 | 34.7 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 1.9 | 7.8 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 1.9 | 15.4 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 1.9 | 11.3 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 1.8 | 5.5 | GO:0070039 | rRNA (guanosine-2'-O-)-methyltransferase activity(GO:0070039) |
| 1.7 | 6.8 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 1.6 | 4.9 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 1.6 | 8.0 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 1.6 | 7.8 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 1.5 | 42.9 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 1.5 | 30.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 1.5 | 9.1 | GO:0070728 | leucine binding(GO:0070728) |
| 1.5 | 7.6 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 1.5 | 12.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 1.5 | 7.4 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 1.4 | 5.6 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 1.4 | 7.0 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 1.4 | 7.0 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 1.4 | 5.6 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 1.4 | 4.1 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 1.3 | 6.4 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 1.2 | 16.9 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 1.2 | 5.9 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 1.1 | 5.6 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 1.1 | 17.9 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 1.1 | 12.9 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 1.1 | 3.2 | GO:0052816 | medium-chain acyl-CoA hydrolase activity(GO:0052815) long-chain acyl-CoA hydrolase activity(GO:0052816) |
| 1.1 | 5.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 1.0 | 7.3 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 1.0 | 5.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 1.0 | 4.0 | GO:0070905 | serine binding(GO:0070905) |
| 1.0 | 8.8 | GO:0043426 | MRF binding(GO:0043426) |
| 0.9 | 5.5 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.9 | 4.3 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.9 | 5.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.9 | 24.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.8 | 3.0 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.8 | 12.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.8 | 11.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.7 | 3.6 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.7 | 4.9 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.7 | 4.2 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.7 | 2.1 | GO:0005055 | laminin receptor activity(GO:0005055) |
| 0.7 | 8.7 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.7 | 4.7 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.7 | 4.6 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.7 | 10.5 | GO:0048185 | activin binding(GO:0048185) |
| 0.6 | 2.6 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.6 | 3.8 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.6 | 8.8 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.6 | 8.9 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.6 | 3.5 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.6 | 4.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.6 | 5.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.5 | 4.9 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.5 | 4.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.5 | 2.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.5 | 3.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.5 | 2.6 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.5 | 5.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.5 | 6.4 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.5 | 2.4 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.5 | 8.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.5 | 9.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.4 | 9.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.4 | 7.0 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.4 | 3.9 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.4 | 8.1 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.4 | 3.8 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.4 | 2.1 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.4 | 7.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.4 | 17.5 | GO:0008066 | glutamate receptor activity(GO:0008066) |
| 0.4 | 4.7 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.4 | 6.6 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.4 | 1.5 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.4 | 8.0 | GO:0005112 | Notch binding(GO:0005112) |
| 0.4 | 1.5 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.4 | 12.1 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.4 | 12.9 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.4 | 2.3 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.4 | 6.2 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.4 | 3.3 | GO:0089720 | caspase binding(GO:0089720) |
| 0.4 | 3.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.4 | 11.7 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.3 | 15.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.3 | 6.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.3 | 35.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.3 | 3.8 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.3 | 4.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.3 | 2.4 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.3 | 1.2 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) scavenger receptor binding(GO:0005124) |
| 0.3 | 2.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.3 | 1.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.3 | 0.9 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.3 | 2.9 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.3 | 2.3 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.3 | 1.4 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.3 | 1.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.3 | 1.9 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.3 | 18.5 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.3 | 7.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.3 | 3.6 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.3 | 1.3 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.3 | 2.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.2 | 1.5 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.2 | 3.0 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.2 | 3.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 12.9 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.2 | 29.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 6.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 26.0 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.2 | 0.7 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.2 | 1.8 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 0.7 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.2 | 6.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 6.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.2 | 3.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 3.9 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 5.1 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.2 | 9.4 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.2 | 3.1 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.2 | 3.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 1.7 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.2 | 3.9 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.2 | 3.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.2 | 12.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 2.2 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.2 | 1.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.2 | 0.4 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.2 | 8.2 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.2 | 28.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 0.9 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.2 | 0.9 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.2 | 1.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 6.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 19.7 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.2 | 9.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 1.6 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.2 | 2.0 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 2.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 1.0 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 5.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 6.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 1.0 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.1 | 1.7 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.1 | 1.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 1.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.4 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.1 | 6.6 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 2.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 18.2 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.1 | 10.3 | GO:0052813 | phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 0.9 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 0.8 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.1 | 7.0 | GO:0030551 | cyclic nucleotide binding(GO:0030551) |
| 0.1 | 0.6 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 8.5 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 7.0 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.1 | 9.7 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 1.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 2.0 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.1 | 4.3 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 1.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 12.8 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 3.2 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 1.0 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 1.1 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 1.4 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.1 | 0.5 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.1 | 5.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 3.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.6 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 5.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 21.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 67.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 0.8 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.1 | 1.0 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 4.5 | GO:1901476 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.1 | 4.1 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 4.1 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 9.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 10.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 3.1 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 6.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 3.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 8.0 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.1 | 3.8 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 1.9 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 2.0 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.7 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 17.5 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.2 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 1.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 2.6 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 2.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.7 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.3 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 1.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.3 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 4.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.8 | 26.8 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.6 | 17.9 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.6 | 4.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.5 | 9.0 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.5 | 20.4 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.5 | 21.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.5 | 9.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.4 | 14.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.4 | 7.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.4 | 27.2 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.4 | 10.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.4 | 15.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.4 | 15.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.3 | 26.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 42.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 12.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 7.1 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.2 | 4.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.2 | 13.9 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.2 | 15.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.2 | 5.3 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.2 | 7.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 7.0 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.2 | 3.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 12.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 12.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 30.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 5.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 4.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 8.3 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 13.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 4.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 14.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 5.4 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.1 | 1.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 1.6 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 4.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 2.3 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 3.0 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 1.2 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 1.5 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.1 | 2.4 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.1 | 0.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 1.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 1.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.2 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 13.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 3.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.6 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.0 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 22.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 1.3 | 21.7 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 1.1 | 21.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 1.0 | 20.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.8 | 64.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.7 | 10.0 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.7 | 1.4 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.7 | 4.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.7 | 9.9 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.6 | 7.4 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.5 | 17.9 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.5 | 3.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.5 | 12.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.5 | 20.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.5 | 22.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.5 | 8.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.5 | 12.5 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.5 | 33.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.5 | 6.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.4 | 0.8 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.4 | 16.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.4 | 7.5 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.4 | 5.5 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.4 | 7.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.3 | 8.0 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.3 | 12.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.3 | 6.4 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.3 | 29.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.3 | 14.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.3 | 11.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.3 | 17.8 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.3 | 16.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.3 | 15.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.3 | 7.7 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.3 | 11.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.3 | 8.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.2 | 32.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.2 | 3.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 4.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 5.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 1.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.2 | 3.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 1.7 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.2 | 4.9 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.2 | 3.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.2 | 9.0 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.2 | 55.8 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.2 | 5.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 10.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 2.8 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.1 | 5.5 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.1 | 3.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.7 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 3.7 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.1 | 2.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 5.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 3.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 2.1 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 3.0 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 7.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 10.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 13.2 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.1 | 1.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 6.9 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 10.3 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 2.1 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.1 | 2.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.2 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 2.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.7 | REACTOME BILE ACID AND BILE SALT METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 1.1 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |