Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for ZBTB7A_ZBTB7C
Z-value: 1.93
Transcription factors associated with ZBTB7A_ZBTB7C
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
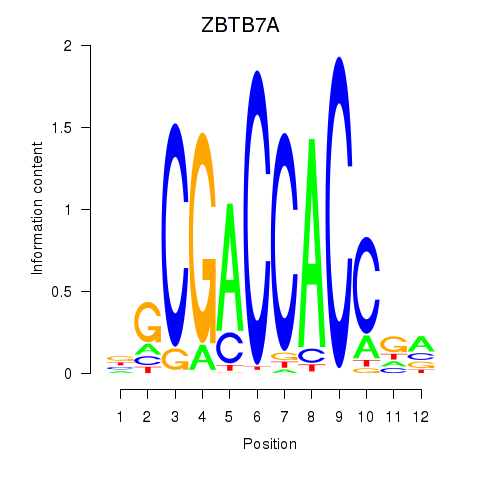
ZBTB7A
|
ENSG00000178951.9 | ZBTB7A |
|
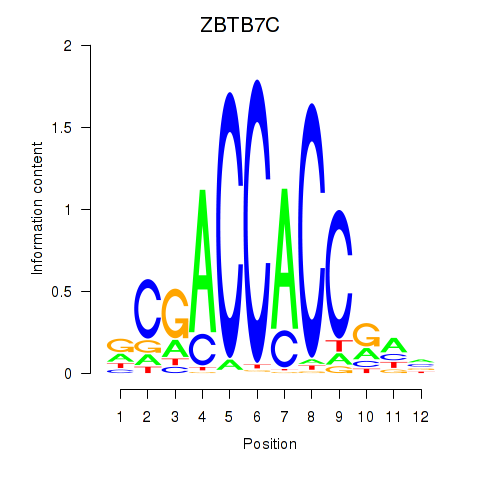
ZBTB7C
|
ENSG00000184828.10 | ZBTB7C |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB7A | hg38_v1_chr19_-_4066892_4066906 | 0.35 | 1.6e-07 | Click! |
| ZBTB7C | hg38_v1_chr18_-_48409292_48409401, hg38_v1_chr18_-_48137295_48137361 | 0.01 | 8.4e-01 | Click! |
Activity profile of ZBTB7A_ZBTB7C motif
Sorted Z-values of ZBTB7A_ZBTB7C motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB7A_ZBTB7C
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.9 | 26.6 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) positive regulation of potassium ion import(GO:1903288) |
| 6.8 | 33.9 | GO:1902613 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 4.1 | 16.4 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 3.9 | 11.7 | GO:0021678 | fourth ventricle development(GO:0021592) third ventricle development(GO:0021678) |
| 3.8 | 19.0 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 3.7 | 18.4 | GO:0007412 | axon target recognition(GO:0007412) |
| 2.7 | 8.2 | GO:0001757 | somite specification(GO:0001757) |
| 2.4 | 14.4 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 1.8 | 8.8 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 1.6 | 14.0 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 1.5 | 4.4 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 1.5 | 4.4 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 1.4 | 7.0 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 1.3 | 11.3 | GO:0009249 | protein lipoylation(GO:0009249) |
| 1.2 | 13.0 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 1.1 | 3.3 | GO:1904907 | negative regulation of maintenance of sister chromatid cohesion(GO:0034092) negative regulation of maintenance of mitotic sister chromatid cohesion(GO:0034183) protein auto-ADP-ribosylation(GO:0070213) maintenance of mitotic sister chromatid cohesion, telomeric(GO:0099403) mitotic sister chromatid cohesion, telomeric(GO:0099404) regulation of telomeric DNA binding(GO:1904742) regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904907) negative regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904908) |
| 1.0 | 3.0 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.9 | 2.8 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.9 | 2.8 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.8 | 10.1 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.8 | 2.5 | GO:0071072 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) negative regulation of phospholipid biosynthetic process(GO:0071072) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.8 | 3.1 | GO:0071110 | protein biotinylation(GO:0009305) histone biotinylation(GO:0071110) |
| 0.8 | 3.0 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.7 | 5.2 | GO:2000768 | positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.7 | 3.0 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.7 | 5.8 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.6 | 9.6 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.6 | 13.4 | GO:0021756 | striatum development(GO:0021756) |
| 0.6 | 47.1 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.5 | 2.1 | GO:2000170 | positive regulation of actin filament-based movement(GO:1903116) positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.5 | 3.8 | GO:1900045 | negative regulation of histone ubiquitination(GO:0033183) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.5 | 22.0 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.5 | 6.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.4 | 4.0 | GO:1905146 | lysosomal protein catabolic process(GO:1905146) |
| 0.3 | 1.7 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.3 | 1.9 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.3 | 1.9 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.3 | 2.5 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.3 | 3.9 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.3 | 1.4 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.3 | 3.3 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.3 | 2.5 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.2 | 0.7 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.2 | 11.2 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.2 | 0.9 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.2 | 3.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.2 | 6.3 | GO:2000505 | regulation of energy homeostasis(GO:2000505) |
| 0.2 | 2.3 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.2 | 1.8 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.2 | 2.5 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.2 | 4.8 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.2 | 3.9 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.2 | 5.9 | GO:0031114 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.2 | 1.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 6.2 | GO:0061641 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.2 | 0.7 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.2 | 6.9 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.2 | 5.7 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.2 | 5.1 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.1 | 4.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 2.2 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.1 | 1.2 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.1 | 2.2 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 3.4 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.1 | 38.8 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.1 | 3.4 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 14.7 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.1 | 1.3 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.1 | 0.7 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 6.1 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.1 | 2.1 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.1 | 1.8 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 1.9 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 0.6 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.1 | GO:2000832 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 | 0.7 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 1.3 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 0.4 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 11.9 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 0.3 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 2.6 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 1.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.5 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 3.0 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 3.9 | GO:0030509 | BMP signaling pathway(GO:0030509) |
| 0.0 | 0.6 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.7 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.0 | 0.6 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.7 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 1.3 | GO:0060071 | Wnt signaling pathway, planar cell polarity pathway(GO:0060071) |
| 0.0 | 0.7 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.8 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.4 | GO:0048536 | spleen development(GO:0048536) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.6 | 61.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 2.3 | 11.7 | GO:0097513 | myosin II filament(GO:0097513) |
| 1.5 | 26.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 1.3 | 4.0 | GO:0097636 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 1.2 | 7.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 1.0 | 2.9 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.9 | 8.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.9 | 4.4 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.8 | 14.0 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.7 | 2.0 | GO:0005606 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) |
| 0.6 | 18.7 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.5 | 19.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.5 | 2.6 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.5 | 12.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.5 | 1.9 | GO:1990393 | 3M complex(GO:1990393) |
| 0.5 | 1.8 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.4 | 2.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.4 | 6.1 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.4 | 3.9 | GO:0070449 | elongin complex(GO:0070449) |
| 0.3 | 6.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.3 | 3.0 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.3 | 4.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.3 | 18.4 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.3 | 26.4 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.3 | 2.8 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.2 | 1.7 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.2 | 8.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.2 | 2.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.2 | 3.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.2 | 3.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.2 | 2.9 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 8.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 3.0 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.1 | 3.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 3.0 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 3.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 3.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 9.4 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.4 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.4 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 2.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 9.0 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 2.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 3.4 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 2.2 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 2.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 2.7 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 1.3 | GO:0036064 | ciliary basal body(GO:0036064) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 19.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 3.1 | 18.4 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 2.9 | 14.4 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 2.0 | 26.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 1.8 | 7.0 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 1.6 | 6.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 1.0 | 8.8 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 1.0 | 2.9 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.8 | 10.1 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.8 | 47.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.8 | 3.1 | GO:0004078 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 0.7 | 5.8 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.6 | 3.0 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.5 | 3.5 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.5 | 3.4 | GO:0070739 | NEDD8 transferase activity(GO:0019788) protein-glutamic acid ligase activity(GO:0070739) |
| 0.5 | 22.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.4 | 2.6 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.4 | 3.9 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.4 | 6.1 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.4 | 3.4 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.4 | 3.8 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.4 | 1.8 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.4 | 12.3 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.3 | 8.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.3 | 1.3 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.3 | 3.0 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.3 | 1.8 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.3 | 2.6 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.3 | 4.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.3 | 9.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.3 | 3.0 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.3 | 9.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.3 | 2.8 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 1.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.2 | 8.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 2.5 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 5.7 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.2 | 0.7 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.2 | 4.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 3.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 5.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 8.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 1.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 4.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 2.8 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 11.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 2.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 1.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 1.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 1.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 13.6 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 1.9 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 1.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.4 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 6.3 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 2.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.7 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 1.8 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 2.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 5.1 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 1.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 2.3 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.6 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 1.0 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.7 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 1.8 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 2.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.6 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 16.0 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.3 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.2 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 1.3 | GO:0017048 | Rho GTPase binding(GO:0017048) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 31.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.4 | 7.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 6.1 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 14.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 2.3 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 4.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 3.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 2.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 1.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 6.7 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 2.9 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 2.0 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 5.1 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.8 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 13.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.5 | 28.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.3 | 13.0 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.3 | 12.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 6.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.3 | 6.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 7.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 9.4 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.2 | 14.3 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.2 | 7.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 4.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 19.0 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 4.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 3.0 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 5.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 2.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 3.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 6.9 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 1.8 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 16.1 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.1 | 6.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 3.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 2.9 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 0.7 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 6.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 1.6 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 9.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 6.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 3.9 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.6 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |