Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for ZIC2_GLI1
Z-value: 9.66
Transcription factors associated with ZIC2_GLI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
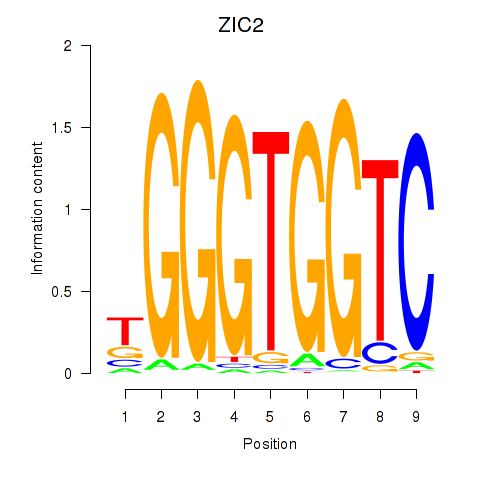
ZIC2
|
ENSG00000043355.12 | ZIC2 |
|
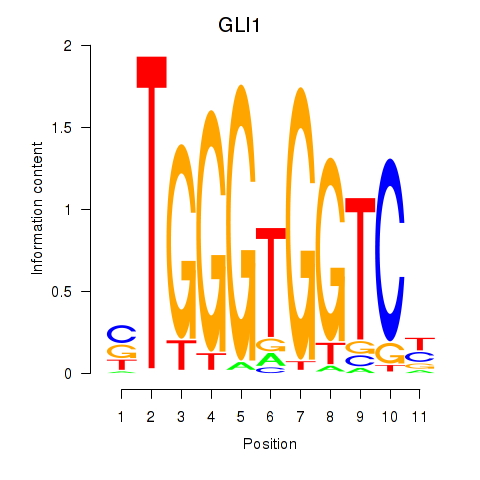
GLI1
|
ENSG00000111087.10 | GLI1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GLI1 | hg38_v1_chr12_+_57459782_57459796, hg38_v1_chr12_+_57460127_57460151 | 0.67 | 9.0e-30 | Click! |
Activity profile of ZIC2_GLI1 motif
Sorted Z-values of ZIC2_GLI1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZIC2_GLI1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.9 | 286.9 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 8.6 | 43.2 | GO:1902613 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 8.5 | 42.3 | GO:0030070 | insulin processing(GO:0030070) |
| 6.3 | 25.1 | GO:1904980 | positive regulation of endosome organization(GO:1904980) |
| 6.1 | 30.7 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 5.8 | 17.3 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 5.6 | 28.1 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 4.8 | 19.4 | GO:0010157 | response to chlorate(GO:0010157) |
| 4.4 | 39.9 | GO:0009249 | protein lipoylation(GO:0009249) |
| 4.0 | 12.0 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 3.9 | 38.5 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 3.8 | 11.3 | GO:0060032 | notochord regression(GO:0060032) |
| 3.6 | 65.7 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 3.5 | 31.9 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 3.5 | 10.5 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 3.2 | 9.5 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 3.1 | 9.4 | GO:0036292 | DNA rewinding(GO:0036292) |
| 3.1 | 12.4 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 2.9 | 14.6 | GO:0090131 | mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) mesenchyme migration(GO:0090131) |
| 2.6 | 20.9 | GO:0060157 | urinary bladder development(GO:0060157) |
| 2.5 | 10.0 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 2.4 | 19.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 2.3 | 9.3 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 2.2 | 6.7 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 2.2 | 59.9 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 2.2 | 24.2 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 2.2 | 8.6 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 2.0 | 6.0 | GO:0019046 | release from viral latency(GO:0019046) |
| 1.9 | 13.6 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 1.9 | 26.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 1.9 | 7.5 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 1.9 | 11.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 1.6 | 14.3 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 1.6 | 7.9 | GO:0072014 | proximal tubule development(GO:0072014) |
| 1.6 | 44.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 1.5 | 13.9 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 1.5 | 35.2 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 1.5 | 20.9 | GO:0007512 | adult heart development(GO:0007512) |
| 1.4 | 10.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 1.4 | 18.3 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 1.4 | 7.0 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 1.3 | 18.6 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 1.2 | 7.1 | GO:1904261 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 1.1 | 7.8 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 1.1 | 39.8 | GO:0061641 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 1.1 | 3.3 | GO:1903570 | coronary vein morphogenesis(GO:0003169) regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 1.0 | 7.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 1.0 | 5.0 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.9 | 11.9 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.9 | 16.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.9 | 5.3 | GO:0035604 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) squamous basal epithelial stem cell differentiation involved in prostate gland acinus development(GO:0060529) |
| 0.9 | 3.4 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 0.8 | 4.9 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.8 | 3.1 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.7 | 12.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.7 | 7.9 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.7 | 2.6 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.6 | 9.0 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.6 | 3.2 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.6 | 14.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.6 | 1.7 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.6 | 7.3 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.5 | 6.5 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.5 | 21.6 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.5 | 4.6 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.5 | 3.4 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.5 | 2.4 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.5 | 1.9 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.5 | 6.5 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.5 | 1.4 | GO:0043697 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.4 | 2.7 | GO:0060482 | lobar bronchus development(GO:0060482) |
| 0.4 | 4.0 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.4 | 1.8 | GO:0060023 | soft palate development(GO:0060023) |
| 0.4 | 5.9 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.4 | 3.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.4 | 4.1 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.4 | 5.2 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.4 | 1.9 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.4 | 7.2 | GO:0060746 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.4 | 3.6 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.3 | 1.4 | GO:1902775 | mitochondrial ribosome assembly(GO:0061668) mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.3 | 9.4 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.3 | 12.9 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.3 | 1.0 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.3 | 6.5 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.3 | 10.3 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.3 | 7.0 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.3 | 0.9 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.3 | 4.7 | GO:0070977 | bone maturation(GO:0070977) |
| 0.3 | 0.5 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.2 | 2.0 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.2 | 2.0 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.2 | 3.8 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.2 | 2.7 | GO:0071285 | urea cycle(GO:0000050) cellular response to lithium ion(GO:0071285) |
| 0.2 | 0.4 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.2 | 2.7 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.2 | 0.7 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.2 | 10.8 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 5.9 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.1 | 8.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 24.6 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.1 | 3.3 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 1.4 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 4.9 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 3.6 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.1 | 2.1 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.1 | 2.0 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.1 | 10.5 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.1 | 1.1 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.1 | 1.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.6 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 2.5 | GO:0050951 | sensory perception of temperature stimulus(GO:0050951) |
| 0.1 | 0.7 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.1 | 4.5 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.1 | 0.8 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.1 | 4.0 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 0.9 | GO:0036315 | cellular response to sterol(GO:0036315) |
| 0.1 | 2.2 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.1 | 21.9 | GO:0032259 | methylation(GO:0032259) |
| 0.1 | 13.2 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.6 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.0 | 0.7 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 1.8 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 10.6 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 1.7 | GO:0002209 | behavioral fear response(GO:0001662) behavioral defense response(GO:0002209) |
| 0.0 | 3.5 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.0 | 0.4 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 1.5 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 2.1 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.2 | GO:0001502 | cartilage condensation(GO:0001502) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.9 | 286.9 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 11.0 | 44.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 4.9 | 14.6 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 3.2 | 16.0 | GO:0036128 | CatSper complex(GO:0036128) |
| 2.9 | 35.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 2.5 | 10.0 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 2.2 | 19.4 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 1.9 | 38.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 1.9 | 31.9 | GO:0033010 | paranodal junction(GO:0033010) |
| 1.8 | 18.2 | GO:0045180 | basal cortex(GO:0045180) |
| 1.5 | 59.9 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 1.5 | 16.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 1.0 | 18.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.9 | 30.1 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.8 | 10.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.8 | 18.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.7 | 12.9 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.6 | 13.6 | GO:0042627 | chylomicron(GO:0042627) |
| 0.6 | 1.8 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.6 | 3.4 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.5 | 14.5 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.5 | 17.0 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.5 | 2.4 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.4 | 6.0 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.4 | 57.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.4 | 13.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.4 | 7.8 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.4 | 6.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.3 | 17.3 | GO:0016235 | aggresome(GO:0016235) |
| 0.3 | 2.0 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.3 | 6.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.3 | 24.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.3 | 10.8 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.3 | 32.2 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.3 | 7.5 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.3 | 15.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.3 | 6.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.2 | 2.0 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.2 | 11.3 | GO:0097546 | ciliary base(GO:0097546) |
| 0.2 | 7.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 18.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.2 | 1.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 36.3 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.2 | 1.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 8.9 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.2 | 6.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 4.1 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 1.9 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 23.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 22.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 4.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.6 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 1.8 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 5.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 3.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 4.0 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 13.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 10.7 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.1 | 1.0 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 4.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 8.6 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 1.6 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 8.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 14.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 3.6 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.5 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 11.8 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.8 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 1.4 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.8 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 2.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 35.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.0 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 4.4 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 5.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 5.1 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 4.8 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.9 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.3 | GO:0002102 | podosome(GO:0002102) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.0 | 44.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 9.8 | 293.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 7.5 | 22.4 | GO:0005055 | laminin receptor activity(GO:0005055) |
| 5.6 | 22.4 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 4.5 | 13.6 | GO:0017129 | triglyceride binding(GO:0017129) |
| 3.6 | 25.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 3.1 | 9.3 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 3.1 | 12.4 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 3.0 | 27.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 2.9 | 47.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 2.7 | 45.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 2.3 | 38.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 2.2 | 8.6 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 2.0 | 7.9 | GO:0035939 | microsatellite binding(GO:0035939) |
| 1.9 | 28.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 1.8 | 14.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 1.5 | 5.9 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 1.4 | 12.8 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 1.3 | 18.8 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 1.3 | 20.9 | GO:0016918 | retinal binding(GO:0016918) retinol binding(GO:0019841) |
| 1.0 | 38.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 1.0 | 23.0 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.9 | 28.2 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.9 | 9.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.9 | 9.4 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.9 | 3.4 | GO:0070905 | serine binding(GO:0070905) |
| 0.8 | 10.0 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.8 | 6.5 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.8 | 14.5 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.8 | 10.0 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.7 | 6.0 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.7 | 6.5 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.7 | 12.0 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.7 | 2.0 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.7 | 3.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.6 | 9.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.6 | 9.5 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.6 | 21.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.6 | 1.7 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.6 | 71.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.5 | 4.9 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.5 | 5.9 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.5 | 3.8 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.5 | 4.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.4 | 11.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.4 | 8.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.4 | 5.8 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.3 | 1.4 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.3 | 4.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.3 | 9.8 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 3.3 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.2 | 4.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.2 | 0.9 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 7.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 4.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 1.9 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 7.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 3.6 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.2 | 1.8 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.2 | 20.4 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.2 | 9.4 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.2 | 34.5 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.2 | 0.8 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.2 | 26.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 19.4 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 10.1 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 3.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 3.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 7.2 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 2.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 4.1 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 0.9 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 7.8 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 10.0 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.1 | 4.1 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.1 | 8.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 4.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 18.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 2.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.9 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 1.5 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 4.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 1.4 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.1 | 19.5 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 1.0 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.1 | 1.9 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 3.4 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 0.7 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 11.9 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 5.3 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 0.6 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 1.8 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.7 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.5 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 2.6 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 20.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 3.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 1.4 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 73.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 1.1 | 17.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.9 | 12.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.8 | 43.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.7 | 22.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.6 | 20.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.4 | 18.6 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.4 | 31.8 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.4 | 14.3 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.3 | 24.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.2 | 49.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 10.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.2 | 55.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 3.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 11.3 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.2 | 2.0 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.2 | 8.8 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 4.0 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 3.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 3.1 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 5.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 3.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 4.2 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 5.0 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 3.5 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 3.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 4.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 3.2 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 1.8 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 4.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 38.5 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 2.7 | 35.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 2.4 | 47.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 1.7 | 39.8 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 1.7 | 42.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 1.6 | 59.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 1.3 | 9.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.8 | 15.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.8 | 5.5 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.6 | 17.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.6 | 13.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.6 | 7.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.5 | 25.1 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.4 | 32.9 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.4 | 16.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 8.6 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.3 | 12.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.3 | 5.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.3 | 14.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.3 | 16.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.2 | 4.7 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.2 | 6.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 4.9 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 3.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 7.3 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.2 | 4.1 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.2 | 12.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 16.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 7.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 7.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 7.9 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 11.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 0.9 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 3.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 6.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 2.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 3.1 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 1.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 4.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 8.9 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 1.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.0 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |