Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
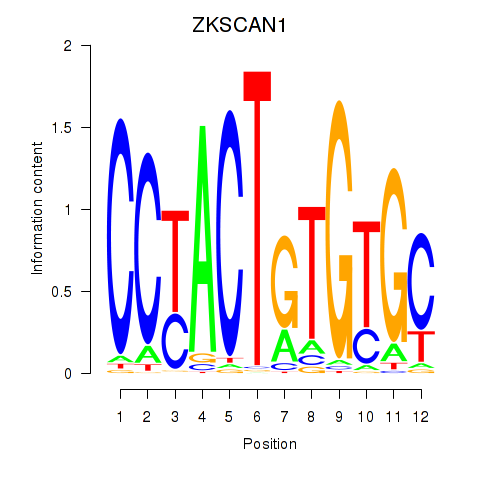
Results for ZKSCAN1
Z-value: 4.77
Transcription factors associated with ZKSCAN1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZKSCAN1
|
ENSG00000106261.17 | ZKSCAN1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZKSCAN1 | hg38_v1_chr7_+_100015588_100015613 | -0.23 | 6.6e-04 | Click! |
Activity profile of ZKSCAN1 motif
Sorted Z-values of ZKSCAN1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZKSCAN1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_136059151 | 54.01 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor beta induced |
| chr5_+_136058849 | 48.31 |
ENST00000508076.5
|
TGFBI
|
transforming growth factor beta induced |
| chr17_-_75154503 | 25.59 |
ENST00000409753.8
ENST00000581874.1 |
JPT1
|
Jupiter microtubule associated homolog 1 |
| chrX_+_47582408 | 21.54 |
ENST00000456754.6
ENST00000218388.9 ENST00000377017.5 ENST00000441738.1 |
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr17_-_75154534 | 21.25 |
ENST00000356033.8
|
JPT1
|
Jupiter microtubule associated homolog 1 |
| chr19_+_38647679 | 19.31 |
ENST00000390009.7
ENST00000589528.1 |
ACTN4
|
actinin alpha 4 |
| chr11_-_58575846 | 18.79 |
ENST00000395074.7
|
LPXN
|
leupaxin |
| chr22_-_37244237 | 16.22 |
ENST00000401529.3
ENST00000249071.11 |
RAC2
|
Rac family small GTPase 2 |
| chrX_-_110440218 | 14.93 |
ENST00000372057.1
ENST00000372054.3 |
AMMECR1
GNG5P2
|
AMMECR nuclear protein 1 G protein subunit gamma 5 pseudogene 2 |
| chr11_+_70203287 | 13.83 |
ENST00000301838.5
|
FADD
|
Fas associated via death domain |
| chr1_-_53238485 | 13.10 |
ENST00000371466.4
ENST00000371470.8 |
MAGOH
|
mago homolog, exon junction complex subunit |
| chr1_+_62436297 | 12.31 |
ENST00000452143.5
ENST00000442679.5 ENST00000371146.5 |
USP1
|
ubiquitin specific peptidase 1 |
| chr15_-_68820861 | 12.13 |
ENST00000560303.1
ENST00000465139.6 |
ANP32A
|
acidic nuclear phosphoprotein 32 family member A |
| chr15_-_34343112 | 11.86 |
ENST00000557912.1
ENST00000328848.6 |
NOP10
|
NOP10 ribonucleoprotein |
| chr15_+_52019206 | 11.68 |
ENST00000681888.1
ENST00000261845.7 ENST00000680066.1 ENST00000680777.1 ENST00000680652.1 |
MAPK6
|
mitogen-activated protein kinase 6 |
| chr5_-_135399863 | 11.59 |
ENST00000510038.1
ENST00000304332.8 |
MACROH2A1
|
macroH2A.1 histone |
| chr19_+_38647614 | 11.44 |
ENST00000252699.7
ENST00000424234.7 ENST00000440400.2 |
ACTN4
|
actinin alpha 4 |
| chr8_-_63038788 | 10.12 |
ENST00000518113.2
ENST00000260118.7 ENST00000677482.1 |
GGH
|
gamma-glutamyl hydrolase |
| chr3_+_184363427 | 10.08 |
ENST00000429568.1
|
POLR2H
|
RNA polymerase II, I and III subunit H |
| chr1_+_65147657 | 9.96 |
ENST00000546702.5
|
AK4
|
adenylate kinase 4 |
| chr3_+_184363387 | 9.91 |
ENST00000452961.5
|
POLR2H
|
RNA polymerase II, I and III subunit H |
| chr22_-_37244417 | 9.83 |
ENST00000405484.5
ENST00000441619.5 ENST00000406508.5 |
RAC2
|
Rac family small GTPase 2 |
| chr19_-_18941184 | 9.04 |
ENST00000594794.5
ENST00000392351.8 ENST00000596482.5 |
HOMER3
|
homer scaffold protein 3 |
| chr17_+_77376083 | 8.99 |
ENST00000427674.6
|
SEPTIN9
|
septin 9 |
| chr3_+_184363351 | 8.65 |
ENST00000443489.5
|
POLR2H
|
RNA polymerase II, I and III subunit H |
| chr5_+_110738983 | 8.59 |
ENST00000355943.8
ENST00000447245.6 |
SLC25A46
|
solute carrier family 25 member 46 |
| chrX_+_110002635 | 8.04 |
ENST00000372072.7
|
TMEM164
|
transmembrane protein 164 |
| chr17_-_42018488 | 7.85 |
ENST00000589773.5
ENST00000674214.1 |
DNAJC7
|
DnaJ heat shock protein family (Hsp40) member C7 |
| chr19_-_18938982 | 7.65 |
ENST00000594439.5
ENST00000221222.15 |
HOMER3
|
homer scaffold protein 3 |
| chr20_-_50153637 | 7.32 |
ENST00000341698.2
ENST00000371652.9 ENST00000371650.9 |
PEDS1-UBE2V1
PEDS1
|
PEDS1-UBE2V1 readthrough plasmanylethanolamine desaturase 1 |
| chr5_-_132227472 | 6.90 |
ENST00000428369.6
|
P4HA2
|
prolyl 4-hydroxylase subunit alpha 2 |
| chr3_+_12287859 | 6.55 |
ENST00000309576.11
ENST00000397015.7 |
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr5_-_135399211 | 6.39 |
ENST00000312469.8
ENST00000423969.6 |
MACROH2A1
|
macroH2A.1 histone |
| chr12_+_8843236 | 6.34 |
ENST00000541459.5
|
A2ML1
|
alpha-2-macroglobulin like 1 |
| chr6_+_36676489 | 6.29 |
ENST00000448526.6
|
CDKN1A
|
cyclin dependent kinase inhibitor 1A |
| chr6_+_36676455 | 6.21 |
ENST00000615513.4
|
CDKN1A
|
cyclin dependent kinase inhibitor 1A |
| chr3_+_12287962 | 6.07 |
ENST00000643197.2
ENST00000644622.2 |
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr20_+_44714853 | 6.03 |
ENST00000372865.4
|
CCN5
|
cellular communication network factor 5 |
| chr8_-_130016622 | 5.89 |
ENST00000518283.5
ENST00000519110.5 |
CYRIB
|
CYFIP related Rac1 interactor B |
| chr10_-_86521737 | 5.57 |
ENST00000298767.10
|
WAPL
|
WAPL cohesin release factor |
| chr12_-_119804472 | 5.44 |
ENST00000678087.1
ENST00000677993.1 |
CIT
|
citron rho-interacting serine/threonine kinase |
| chr20_+_44714835 | 5.41 |
ENST00000372868.6
|
CCN5
|
cellular communication network factor 5 |
| chr7_+_74174323 | 5.26 |
ENST00000677570.1
ENST00000265753.13 ENST00000353999.6 ENST00000679266.1 ENST00000679287.1 ENST00000677681.1 |
EIF4H
|
eukaryotic translation initiation factor 4H |
| chr17_+_28744034 | 5.25 |
ENST00000444415.7
ENST00000262396.10 |
TRAF4
|
TNF receptor associated factor 4 |
| chr12_-_119804298 | 5.17 |
ENST00000678652.1
ENST00000678494.1 |
CIT
|
citron rho-interacting serine/threonine kinase |
| chr1_-_36149450 | 5.17 |
ENST00000373163.5
|
TRAPPC3
|
trafficking protein particle complex 3 |
| chr3_+_12287899 | 5.09 |
ENST00000643888.2
|
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr19_-_55149193 | 5.07 |
ENST00000587758.5
ENST00000588981.6 ENST00000356783.9 ENST00000291901.12 ENST00000588426.5 ENST00000536926.5 ENST00000588147.5 |
TNNT1
|
troponin T1, slow skeletal type |
| chr12_+_121888751 | 4.96 |
ENST00000261817.6
ENST00000538613.5 ENST00000542602.1 ENST00000541212.6 |
PSMD9
|
proteasome 26S subunit, non-ATPase 9 |
| chr12_-_48925483 | 4.74 |
ENST00000550765.6
ENST00000552878.5 ENST00000453172.2 |
FKBP11
|
FKBP prolyl isomerase 11 |
| chr19_-_3063101 | 4.57 |
ENST00000221561.12
|
TLE5
|
TLE family member 5, transcriptional modulator |
| chr1_+_65147514 | 4.36 |
ENST00000545314.5
|
AK4
|
adenylate kinase 4 |
| chr6_+_32976347 | 4.34 |
ENST00000606059.1
|
BRD2
|
bromodomain containing 2 |
| chr10_+_110919595 | 4.30 |
ENST00000369452.9
|
SHOC2
|
SHOC2 leucine rich repeat scaffold protein |
| chr22_+_36863091 | 4.04 |
ENST00000650698.1
|
NCF4
|
neutrophil cytosolic factor 4 |
| chr1_+_236795254 | 3.90 |
ENST00000366577.10
ENST00000674797.2 |
MTR
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr1_-_36149464 | 3.89 |
ENST00000373166.8
ENST00000373159.1 ENST00000373162.5 ENST00000616074.4 ENST00000616395.4 |
TRAPPC3
|
trafficking protein particle complex 3 |
| chr6_-_30672984 | 3.03 |
ENST00000415603.1
ENST00000376442.8 |
DHX16
|
DEAH-box helicase 16 |
| chr10_-_110918934 | 2.95 |
ENST00000605742.5
|
BBIP1
|
BBSome interacting protein 1 |
| chr12_-_24902243 | 2.91 |
ENST00000538118.5
|
BCAT1
|
branched chain amino acid transaminase 1 |
| chr16_+_8642375 | 2.86 |
ENST00000562973.1
|
METTL22
|
methyltransferase like 22 |
| chr17_+_28744002 | 2.85 |
ENST00000618771.1
ENST00000262395.10 ENST00000422344.5 |
TRAF4
|
TNF receptor associated factor 4 |
| chr14_-_106639589 | 2.66 |
ENST00000390630.3
|
IGHV4-61
|
immunoglobulin heavy variable 4-61 |
| chr9_-_109498251 | 2.62 |
ENST00000374541.4
ENST00000262539.7 |
PTPN3
|
protein tyrosine phosphatase non-receptor type 3 |
| chr16_+_32066065 | 2.57 |
ENST00000354689.6
|
IGHV3OR16-9
|
immunoglobulin heavy variable 3/OR16-9 (non-functional) |
| chr2_-_74507664 | 2.49 |
ENST00000233630.11
|
PCGF1
|
polycomb group ring finger 1 |
| chr1_+_16022004 | 2.46 |
ENST00000439316.6
|
CLCNKA
|
chloride voltage-gated channel Ka |
| chr22_+_21567705 | 2.45 |
ENST00000342192.9
|
UBE2L3
|
ubiquitin conjugating enzyme E2 L3 |
| chr2_-_65432591 | 2.39 |
ENST00000356388.9
|
SPRED2
|
sprouty related EVH1 domain containing 2 |
| chr15_-_37098281 | 2.26 |
ENST00000559085.5
ENST00000397624.7 |
MEIS2
|
Meis homeobox 2 |
| chrX_-_40177213 | 2.07 |
ENST00000672922.2
ENST00000378455.8 ENST00000342274.8 |
BCOR
|
BCL6 corepressor |
| chr1_+_16022030 | 2.04 |
ENST00000331433.5
|
CLCNKA
|
chloride voltage-gated channel Ka |
| chr17_-_41034871 | 1.97 |
ENST00000344363.7
|
KRTAP1-3
|
keratin associated protein 1-3 |
| chr1_-_149936324 | 1.93 |
ENST00000369140.7
|
MTMR11
|
myotubularin related protein 11 |
| chr15_-_78077657 | 1.90 |
ENST00000300584.8
ENST00000409931.7 |
TBC1D2B
|
TBC1 domain family member 2B |
| chr15_-_19988117 | 1.85 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr6_+_159761991 | 1.78 |
ENST00000367048.5
|
ACAT2
|
acetyl-CoA acetyltransferase 2 |
| chr3_-_52056552 | 1.75 |
ENST00000495880.2
|
DUSP7
|
dual specificity phosphatase 7 |
| chr19_+_51127030 | 1.70 |
ENST00000599948.1
|
SIGLEC9
|
sialic acid binding Ig like lectin 9 |
| chr12_+_112978386 | 1.63 |
ENST00000342315.8
|
OAS2
|
2'-5'-oligoadenylate synthetase 2 |
| chr19_-_3062464 | 1.53 |
ENST00000327141.9
|
TLE5
|
TLE family member 5, transcriptional modulator |
| chr1_+_16021871 | 1.51 |
ENST00000375692.5
|
CLCNKA
|
chloride voltage-gated channel Ka |
| chr22_+_20507571 | 1.43 |
ENST00000414658.5
ENST00000432052.5 ENST00000292733.11 ENST00000263205.11 ENST00000406969.5 |
MED15
|
mediator complex subunit 15 |
| chr15_+_75201873 | 1.37 |
ENST00000394987.5
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr15_+_40439721 | 1.32 |
ENST00000561234.5
|
BAHD1
|
bromo adjacent homology domain containing 1 |
| chr7_-_27130733 | 1.24 |
ENST00000428284.2
ENST00000360046.10 ENST00000610970.1 |
HOXA4
|
homeobox A4 |
| chr19_-_3062772 | 1.12 |
ENST00000586742.5
|
TLE5
|
TLE family member 5, transcriptional modulator |
| chr15_-_79923084 | 1.01 |
ENST00000615374.5
|
ST20-MTHFS
|
ST20-MTHFS readthrough |
| chr10_+_23095556 | 1.00 |
ENST00000376510.8
|
MSRB2
|
methionine sulfoxide reductase B2 |
| chr10_-_101776104 | 0.99 |
ENST00000320185.7
ENST00000344255.8 |
FGF8
|
fibroblast growth factor 8 |
| chr11_-_111379268 | 0.99 |
ENST00000393067.8
|
POU2AF1
|
POU class 2 homeobox associating factor 1 |
| chr16_+_56865202 | 0.93 |
ENST00000566786.5
ENST00000438926.6 ENST00000563236.6 ENST00000262502.5 |
SLC12A3
|
solute carrier family 12 member 3 |
| chr19_+_48019726 | 0.92 |
ENST00000593413.1
|
ELSPBP1
|
epididymal sperm binding protein 1 |
| chr20_-_63568074 | 0.74 |
ENST00000427522.6
|
HELZ2
|
helicase with zinc finger 2 |
| chr2_+_129979641 | 0.68 |
ENST00000410061.4
|
RAB6C
|
RAB6C, member RAS oncogene family |
| chr1_-_151831768 | 0.67 |
ENST00000318247.7
|
RORC
|
RAR related orphan receptor C |
| chr14_+_54509885 | 0.65 |
ENST00000557317.1
ENST00000216420.12 |
CGRRF1
|
cell growth regulator with ring finger domain 1 |
| chr15_+_75206398 | 0.60 |
ENST00000565074.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr5_+_156326735 | 0.54 |
ENST00000435422.7
|
SGCD
|
sarcoglycan delta |
| chr12_-_102481744 | 0.45 |
ENST00000644491.1
|
IGF1
|
insulin like growth factor 1 |
| chr7_-_27130182 | 0.41 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr1_+_16043776 | 0.39 |
ENST00000375679.9
|
CLCNKB
|
chloride voltage-gated channel Kb |
| chr2_-_131364168 | 0.37 |
ENST00000623617.3
|
RAB6D
|
RAB6D, member RAS oncogene family |
| chr6_+_35297809 | 0.30 |
ENST00000316637.7
|
DEF6
|
DEF6 guanine nucleotide exchange factor |
| chr19_+_13024917 | 0.25 |
ENST00000587260.1
|
NFIX
|
nuclear factor I X |
| chr5_+_174724549 | 0.20 |
ENST00000239243.7
ENST00000507785.2 |
MSX2
|
msh homeobox 2 |
| chr1_+_6034980 | 0.16 |
ENST00000378092.6
ENST00000472700.7 |
KCNAB2
|
potassium voltage-gated channel subfamily A regulatory beta subunit 2 |
| chr16_-_11276473 | 0.15 |
ENST00000241808.9
ENST00000435245.2 |
PRM2
|
protamine 2 |
| chr20_-_3663399 | 0.15 |
ENST00000290417.7
ENST00000319242.8 |
GFRA4
|
GDNF family receptor alpha 4 |
| chr3_-_45796467 | 0.08 |
ENST00000353278.8
ENST00000456124.6 |
SLC6A20
|
solute carrier family 6 member 20 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 30.7 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 4.0 | 11.9 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 3.6 | 21.5 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 3.5 | 13.8 | GO:0072683 | T cell extravasation(GO:0072683) |
| 2.9 | 8.6 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 2.5 | 17.7 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 2.5 | 10.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 2.3 | 18.8 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 2.3 | 25.5 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 1.9 | 5.6 | GO:0060623 | establishment of sister chromatid cohesion(GO:0034085) regulation of chromosome condensation(GO:0060623) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 1.7 | 5.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 1.6 | 14.3 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 1.4 | 12.5 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.8 | 8.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.8 | 12.3 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.7 | 28.6 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.6 | 102.2 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.6 | 16.7 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.6 | 2.9 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.6 | 5.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.5 | 5.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.5 | 2.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.5 | 1.0 | GO:0060128 | corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.5 | 7.9 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.3 | 6.9 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.3 | 2.6 | GO:0098902 | regulation of membrane depolarization during action potential(GO:0098902) |
| 0.3 | 3.9 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.2 | 12.1 | GO:0043486 | histone exchange(GO:0043486) |
| 0.2 | 10.6 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.2 | 2.4 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.2 | 13.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.2 | 1.5 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.1 | 2.5 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.1 | 1.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 1.3 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 4.0 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.1 | 9.1 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 7.1 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 5.2 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.1 | 0.4 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.1 | 0.9 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.1 | 4.3 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 4.3 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.1 | 2.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.9 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 1.7 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.7 | GO:0060850 | regulation of transcription involved in cell fate commitment(GO:0060850) |
| 0.0 | 2.3 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.3 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 1.9 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.2 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 2.9 | GO:0018022 | peptidyl-lysine methylation(GO:0018022) |
| 0.0 | 1.8 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.7 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 2.0 | GO:0031424 | keratinization(GO:0031424) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 12.5 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 1.8 | 28.6 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 1.7 | 11.9 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 1.0 | 30.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.9 | 120.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.8 | 9.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.7 | 13.8 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.6 | 5.6 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.5 | 13.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.4 | 4.0 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.4 | 18.8 | GO:0002102 | podosome(GO:0002102) |
| 0.3 | 26.0 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.3 | 5.0 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.3 | 5.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.3 | 4.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 5.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 16.7 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.2 | 1.5 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 7.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 10.1 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 1.3 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 41.3 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 18.7 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 10.6 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.1 | 4.6 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 5.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 0.4 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 8.1 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 2.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 8.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) organelle outer membrane(GO:0031968) |
| 0.0 | 13.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 5.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.4 | GO:0030658 | transport vesicle membrane(GO:0030658) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 14.3 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 2.5 | 12.5 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 1.8 | 28.6 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 1.8 | 17.7 | GO:0050692 | DBD domain binding(GO:0050692) |
| 1.7 | 11.9 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 1.5 | 13.8 | GO:0089720 | caspase binding(GO:0089720) |
| 1.4 | 6.9 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 1.2 | 102.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.9 | 18.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.9 | 16.7 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.8 | 2.4 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.7 | 30.7 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.6 | 2.9 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.6 | 3.9 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.5 | 4.0 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.5 | 10.1 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.4 | 8.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.4 | 1.8 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.4 | 5.3 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.4 | 11.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.4 | 5.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.2 | 5.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 7.9 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.2 | 2.5 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.2 | 1.0 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.2 | 9.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 2.0 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 12.3 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 7.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 1.7 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 4.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.7 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 4.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.3 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.1 | 9.1 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 26.0 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.1 | 0.9 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 12.1 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 0.9 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.1 | 2.6 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 3.0 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 31.4 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.0 | 2.9 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 1.0 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.2 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.2 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 1.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 1.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 102.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.9 | 12.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.4 | 21.9 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.3 | 17.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.3 | 13.8 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.3 | 12.3 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.2 | 19.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.2 | 30.7 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.2 | 10.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 12.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 8.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 2.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 2.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 4.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 2.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.4 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 1.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 102.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 1.3 | 26.8 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 1.0 | 12.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.8 | 30.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.7 | 13.8 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.6 | 26.0 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.6 | 12.5 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.5 | 21.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 13.1 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.2 | 18.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 3.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 17.1 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.1 | 2.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 4.0 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.1 | 1.7 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 5.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 5.3 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.1 | 1.0 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 1.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 2.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |