Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
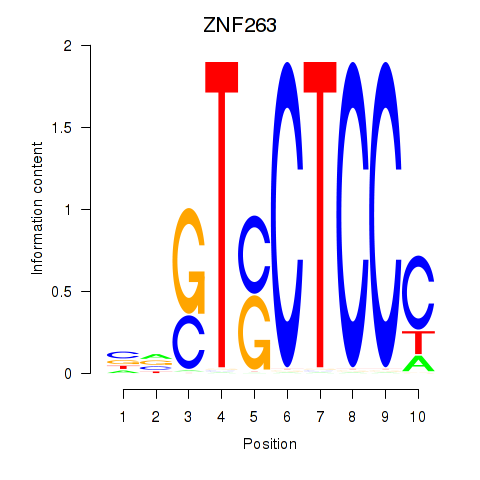
Results for ZNF263
Z-value: 6.96
Transcription factors associated with ZNF263
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF263
|
ENSG00000006194.10 | ZNF263 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF263 | hg38_v1_chr16_+_3283443_3283544 | -0.40 | 5.5e-10 | Click! |
Activity profile of ZNF263 motif
Sorted Z-values of ZNF263 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF263
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.8 | 17.6 | GO:0035793 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 6.8 | 20.3 | GO:0098759 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) interleukin-1 beta biosynthetic process(GO:0050720) response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) regulation of progesterone biosynthetic process(GO:2000182) |
| 6.2 | 18.7 | GO:0036166 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 4.8 | 14.3 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 4.5 | 22.4 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 4.4 | 17.4 | GO:0033594 | response to hydroxyisoflavone(GO:0033594) |
| 4.1 | 12.2 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 4.0 | 52.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 3.8 | 15.2 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 3.7 | 14.6 | GO:1903116 | positive regulation of actin filament-based movement(GO:1903116) positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 3.5 | 10.6 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 3.5 | 10.6 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 3.5 | 3.5 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 3.3 | 10.0 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) cell proliferation involved in heart valve development(GO:2000793) |
| 3.3 | 13.1 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 3.1 | 33.7 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 3.0 | 12.0 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) growth plate cartilage chondrocyte development(GO:0003431) |
| 3.0 | 3.0 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 2.9 | 14.5 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 2.9 | 20.1 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 2.9 | 8.6 | GO:0043465 | fermentation(GO:0006113) regulation of fermentation(GO:0043465) |
| 2.9 | 11.4 | GO:0061443 | endocardial cushion cell differentiation(GO:0061443) |
| 2.8 | 8.4 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 2.7 | 10.9 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 2.6 | 5.2 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 2.6 | 33.2 | GO:0045794 | response to carbon monoxide(GO:0034465) negative regulation of cell volume(GO:0045794) |
| 2.4 | 7.3 | GO:0071464 | cellular response to hydrostatic pressure(GO:0071464) |
| 2.4 | 2.4 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 2.4 | 21.6 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 2.3 | 7.0 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 2.3 | 7.0 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 2.3 | 7.0 | GO:1900920 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 2.3 | 6.9 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 2.2 | 8.7 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 2.2 | 6.5 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) mast cell proliferation(GO:0070662) |
| 2.1 | 10.3 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 2.0 | 6.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 2.0 | 2.0 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 1.9 | 5.7 | GO:0002265 | astrocyte activation involved in immune response(GO:0002265) |
| 1.9 | 7.6 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 1.9 | 9.4 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 1.9 | 11.2 | GO:0071873 | response to norepinephrine(GO:0071873) cellular response to norepinephrine stimulus(GO:0071874) |
| 1.8 | 5.4 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 1.8 | 5.4 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 1.8 | 5.3 | GO:0002865 | negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) |
| 1.8 | 5.3 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 1.7 | 10.4 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 1.7 | 19.1 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 1.7 | 5.2 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 1.7 | 10.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 1.7 | 11.8 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 1.7 | 14.9 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 1.6 | 6.6 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 1.6 | 24.0 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 1.6 | 3.2 | GO:0090274 | regulation of somatostatin secretion(GO:0090273) positive regulation of somatostatin secretion(GO:0090274) |
| 1.6 | 4.7 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 1.6 | 7.8 | GO:0044856 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 1.6 | 20.3 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 1.6 | 18.7 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 1.6 | 4.7 | GO:0072237 | metanephric proximal tubule development(GO:0072237) |
| 1.6 | 10.9 | GO:1902856 | negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 1.5 | 4.6 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 1.5 | 21.5 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 1.5 | 4.6 | GO:0060366 | subpallium cell proliferation in forebrain(GO:0022012) lateral ganglionic eminence cell proliferation(GO:0022018) lambdoid suture morphogenesis(GO:0060366) sagittal suture morphogenesis(GO:0060367) anterior semicircular canal development(GO:0060873) lateral semicircular canal development(GO:0060875) |
| 1.5 | 10.7 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 1.5 | 4.4 | GO:0072134 | nephrogenic mesenchyme morphogenesis(GO:0072134) |
| 1.5 | 4.4 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 1.4 | 18.7 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 1.4 | 5.7 | GO:0035625 | negative regulation of epinephrine secretion(GO:0032811) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 1.4 | 4.2 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 1.4 | 4.1 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 1.4 | 6.8 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 1.3 | 25.4 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 1.3 | 3.9 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 1.3 | 3.9 | GO:0060214 | endocardium formation(GO:0060214) |
| 1.3 | 5.1 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 1.3 | 2.5 | GO:0001555 | oocyte growth(GO:0001555) |
| 1.3 | 12.6 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 1.2 | 3.7 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 1.2 | 4.8 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 1.2 | 17.9 | GO:1901978 | positive regulation of cell cycle checkpoint(GO:1901978) |
| 1.2 | 4.7 | GO:0072717 | traversing start control point of mitotic cell cycle(GO:0007089) transcription factor catabolic process(GO:0036369) cellular response to actinomycin D(GO:0072717) |
| 1.2 | 3.5 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 1.2 | 4.6 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 1.1 | 2.3 | GO:1902022 | lysine transport(GO:0015819) lysine import(GO:0034226) L-lysine import(GO:0061461) L-lysine transport(GO:1902022) L-lysine import into cell(GO:1903410) |
| 1.1 | 3.4 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 1.1 | 7.9 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 1.1 | 4.5 | GO:0033504 | floor plate development(GO:0033504) |
| 1.1 | 7.9 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 1.1 | 3.4 | GO:1903984 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 1.1 | 3.4 | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 1.1 | 4.5 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 1.1 | 3.3 | GO:1901963 | regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) |
| 1.1 | 5.4 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 1.1 | 4.3 | GO:0061358 | negative regulation of Wnt protein secretion(GO:0061358) |
| 1.1 | 4.3 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 1.1 | 6.4 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 1.1 | 14.9 | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) |
| 1.1 | 4.2 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 1.1 | 3.2 | GO:0021722 | pons maturation(GO:0021586) superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 1.0 | 4.2 | GO:0060938 | cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 1.0 | 5.2 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) chorionic trophoblast cell differentiation(GO:0060718) |
| 1.0 | 7.2 | GO:0086100 | endothelin receptor signaling pathway(GO:0086100) |
| 1.0 | 3.0 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 1.0 | 5.9 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 1.0 | 3.0 | GO:0007538 | primary sex determination(GO:0007538) |
| 1.0 | 2.0 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 1.0 | 3.9 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 1.0 | 3.9 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 1.0 | 3.8 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 1.0 | 3.8 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.9 | 11.0 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.9 | 2.7 | GO:0090403 | negative regulation of icosanoid secretion(GO:0032304) oxidative stress-induced premature senescence(GO:0090403) negative regulation of phospholipase A2 activity(GO:1900138) |
| 0.9 | 4.5 | GO:0010046 | response to mycotoxin(GO:0010046) iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.9 | 5.3 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.9 | 3.5 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.9 | 2.6 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.9 | 1.7 | GO:1902866 | regulation of neural retina development(GO:0061074) regulation of retina development in camera-type eye(GO:1902866) |
| 0.9 | 2.6 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.9 | 6.0 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.9 | 6.0 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.9 | 6.0 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.8 | 2.5 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.8 | 3.4 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.8 | 3.3 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.8 | 2.5 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.8 | 4.9 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) determination of liver left/right asymmetry(GO:0071910) |
| 0.8 | 3.3 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.8 | 1.6 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.8 | 19.6 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.8 | 5.7 | GO:0043586 | tongue development(GO:0043586) |
| 0.8 | 4.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.8 | 4.0 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.8 | 4.0 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.8 | 2.4 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.8 | 12.8 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.8 | 3.2 | GO:0032903 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.8 | 9.5 | GO:0097012 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.8 | 1.6 | GO:0048852 | hypophysis morphogenesis(GO:0048850) diencephalon morphogenesis(GO:0048852) |
| 0.8 | 6.2 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.8 | 3.8 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.8 | 3.1 | GO:1902732 | condensed mesenchymal cell proliferation(GO:0072137) positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.8 | 3.8 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.8 | 2.3 | GO:0021637 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.7 | 3.0 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.7 | 1.5 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.7 | 5.2 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.7 | 2.9 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.7 | 1.4 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.7 | 4.2 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.7 | 11.9 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.7 | 2.1 | GO:0034471 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.7 | 6.2 | GO:0000101 | sulfur amino acid transport(GO:0000101) |
| 0.7 | 4.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.7 | 2.7 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.7 | 13.5 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.7 | 2.0 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.7 | 2.0 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.6 | 1.9 | GO:0090298 | negative regulation of mitochondrial DNA replication(GO:0090298) negative regulation of mitochondrial DNA metabolic process(GO:1901859) |
| 0.6 | 2.6 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.6 | 3.8 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.6 | 3.8 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.6 | 12.6 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.6 | 6.3 | GO:0032594 | protein transport within lipid bilayer(GO:0032594) |
| 0.6 | 17.3 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.6 | 1.2 | GO:0001806 | type IV hypersensitivity(GO:0001806) |
| 0.6 | 7.3 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.6 | 9.7 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.6 | 19.1 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.6 | 7.0 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.6 | 4.6 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.6 | 2.3 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.6 | 2.9 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.6 | 5.7 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.6 | 2.8 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.6 | 2.8 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.6 | 3.9 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.5 | 5.8 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.5 | 2.1 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.5 | 4.2 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.5 | 2.6 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.5 | 9.0 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.5 | 4.2 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.5 | 1.6 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.5 | 1.5 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.5 | 8.7 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.5 | 3.0 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.5 | 5.5 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.5 | 4.5 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.5 | 8.9 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.5 | 2.0 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.5 | 1.0 | GO:1904640 | response to methionine(GO:1904640) |
| 0.5 | 3.4 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.5 | 2.4 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.5 | 4.3 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.5 | 1.9 | GO:0090270 | fibroblast growth factor production(GO:0090269) regulation of fibroblast growth factor production(GO:0090270) |
| 0.5 | 3.8 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.5 | 7.0 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.5 | 11.4 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.4 | 3.1 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.4 | 5.3 | GO:0021794 | thalamus development(GO:0021794) |
| 0.4 | 4.0 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.4 | 4.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.4 | 1.8 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.4 | 1.7 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.4 | 2.6 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.4 | 1.3 | GO:0090427 | egg activation(GO:0007343) activation of meiosis(GO:0090427) |
| 0.4 | 2.6 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.4 | 0.9 | GO:0061110 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.4 | 2.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.4 | 0.9 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.4 | 6.7 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.4 | 5.0 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.4 | 6.2 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.4 | 2.5 | GO:1902949 | positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.4 | 5.8 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.4 | 2.9 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) |
| 0.4 | 2.4 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.4 | 3.6 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.4 | 5.7 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.4 | 2.4 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.4 | 6.4 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.4 | 7.5 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.4 | 7.5 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.4 | 5.5 | GO:1904353 | regulation of telomere capping(GO:1904353) |
| 0.4 | 2.4 | GO:0007442 | hindgut morphogenesis(GO:0007442) embryonic hindgut morphogenesis(GO:0048619) hindgut development(GO:0061525) |
| 0.4 | 4.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.4 | 5.1 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.4 | 1.2 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.4 | 3.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.4 | 2.3 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.4 | 4.6 | GO:0035878 | nail development(GO:0035878) |
| 0.4 | 4.2 | GO:0014010 | Schwann cell proliferation(GO:0014010) |
| 0.4 | 7.6 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.4 | 1.1 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.4 | 3.7 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.4 | 6.2 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.4 | 2.6 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.4 | 2.9 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.4 | 6.9 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.4 | 2.9 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.4 | 2.9 | GO:0032960 | regulation of inositol trisphosphate biosynthetic process(GO:0032960) positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.4 | 1.8 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.4 | 1.4 | GO:0032242 | regulation of nucleoside transport(GO:0032242) |
| 0.4 | 6.4 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.4 | 1.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.4 | 5.7 | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.4 | 4.9 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.3 | 7.3 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.3 | 1.7 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.3 | 4.9 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.3 | 9.7 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.3 | 2.4 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.3 | 20.6 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.3 | 4.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.3 | 6.5 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.3 | 2.4 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.3 | 1.0 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.3 | 3.7 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.3 | 2.3 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.3 | 6.0 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.3 | 2.3 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.3 | 2.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.3 | 1.3 | GO:0009822 | alkaloid catabolic process(GO:0009822) isoquinoline alkaloid metabolic process(GO:0033076) |
| 0.3 | 46.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.3 | 1.9 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.3 | 2.6 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.3 | 1.9 | GO:0009624 | response to nematode(GO:0009624) |
| 0.3 | 3.5 | GO:0001696 | gastric acid secretion(GO:0001696) |
| 0.3 | 1.9 | GO:0060529 | squamous basal epithelial stem cell differentiation involved in prostate gland acinus development(GO:0060529) |
| 0.3 | 7.9 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.3 | 1.9 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.3 | 3.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.3 | 3.7 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.3 | 2.5 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.3 | 0.9 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.3 | 5.5 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.3 | 2.4 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 0.3 | 23.2 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.3 | 1.8 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.3 | 1.5 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.3 | 3.9 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.3 | 1.5 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.3 | 4.7 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.3 | 0.9 | GO:0019075 | virus maturation(GO:0019075) |
| 0.3 | 4.6 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.3 | 5.3 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.3 | 1.1 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.3 | 6.6 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.3 | 3.5 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.3 | 9.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.3 | 5.2 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.3 | 1.8 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.3 | 1.3 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.3 | 1.6 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.3 | 2.1 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.3 | 8.0 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.3 | 1.3 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.3 | 14.6 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.3 | 0.8 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.3 | 1.0 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.3 | 1.0 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.3 | 1.8 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.3 | 5.0 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.2 | 1.7 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 1.5 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 3.4 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.2 | 3.4 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.2 | 1.7 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.2 | 6.5 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.2 | 1.7 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.2 | 1.0 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.2 | 4.0 | GO:0033622 | integrin activation(GO:0033622) |
| 0.2 | 0.7 | GO:1900619 | acetylcholine metabolic process(GO:0008291) otolith development(GO:0048840) sensory system development(GO:0048880) acetylcholine secretion(GO:0061526) acetate ester metabolic process(GO:1900619) |
| 0.2 | 4.0 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.2 | 2.6 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.2 | 15.8 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.2 | 3.7 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.2 | 0.9 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.2 | 1.6 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.2 | 2.0 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.2 | 8.5 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.2 | 7.2 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.2 | 0.7 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.2 | 1.8 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.2 | 1.5 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.2 | 0.9 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.2 | 8.1 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.2 | 0.7 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.2 | 5.2 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.2 | 3.0 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.2 | 3.6 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.2 | 0.8 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.2 | 0.8 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 0.2 | 3.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.2 | 4.8 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 0.6 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.2 | 3.4 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.2 | 1.4 | GO:0023019 | signal transduction involved in regulation of gene expression(GO:0023019) |
| 0.2 | 1.7 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.2 | 1.0 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.2 | 0.7 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.2 | 3.5 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.2 | 22.7 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.2 | 0.7 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.2 | 0.4 | GO:0070101 | detection of peptidoglycan(GO:0032499) positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.2 | 4.3 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.2 | 1.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.2 | 2.7 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.2 | 1.6 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.2 | 0.5 | GO:0021860 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.2 | 5.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.2 | 2.6 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.2 | 1.0 | GO:1903284 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) regulation of peroxidase activity(GO:2000468) positive regulation of peroxidase activity(GO:2000470) |
| 0.2 | 3.2 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 2.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.2 | 3.7 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.2 | 2.2 | GO:1903392 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of adherens junction organization(GO:1903392) |
| 0.2 | 1.0 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.2 | 3.1 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.2 | 4.5 | GO:0048806 | genitalia development(GO:0048806) |
| 0.2 | 2.4 | GO:0051412 | response to corticosterone(GO:0051412) |
| 0.2 | 5.9 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.2 | 2.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.2 | 7.3 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.2 | 0.9 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.1 | 1.2 | GO:0003360 | brainstem development(GO:0003360) |
| 0.1 | 3.1 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.1 | 4.7 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.1 | 1.3 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.1 | 1.3 | GO:0045837 | negative regulation of mitochondrial membrane potential(GO:0010917) negative regulation of membrane potential(GO:0045837) |
| 0.1 | 1.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 3.8 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.8 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.1 | 2.0 | GO:0035904 | aorta development(GO:0035904) |
| 0.1 | 2.5 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.1 | 2.3 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 3.3 | GO:0034776 | response to histamine(GO:0034776) cellular response to histamine(GO:0071420) |
| 0.1 | 1.3 | GO:0098907 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.1 | 0.5 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.1 | 0.8 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 0.1 | 4.2 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 4.2 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.1 | 8.1 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 3.2 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 2.6 | GO:0010922 | positive regulation of phosphatase activity(GO:0010922) |
| 0.1 | 5.4 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.1 | 3.0 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 4.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 2.1 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.1 | 1.4 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.1 | 1.5 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 2.9 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.1 | 1.0 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 1.9 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 1.3 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.1 | 1.5 | GO:0098856 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.1 | 0.7 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.1 | 1.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 7.6 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 | 2.1 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.1 | 3.1 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.1 | 2.1 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 0.6 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 4.7 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.1 | 3.8 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.1 | 0.8 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.1 | 4.0 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 1.3 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 6.7 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.1 | 2.2 | GO:0019229 | regulation of vasoconstriction(GO:0019229) |
| 0.1 | 2.2 | GO:0001947 | heart looping(GO:0001947) |
| 0.1 | 0.6 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.1 | 0.5 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 0.3 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.1 | 1.4 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 5.6 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 1.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 14.9 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.1 | 5.8 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 2.1 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 0.9 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.1 | 1.6 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.1 | 0.9 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 2.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.9 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.1 | 1.1 | GO:0072678 | T cell migration(GO:0072678) |
| 0.1 | 3.3 | GO:0042472 | inner ear morphogenesis(GO:0042472) |
| 0.1 | 0.9 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.1 | 1.9 | GO:0071305 | cellular response to vitamin D(GO:0071305) |
| 0.1 | 8.8 | GO:0007626 | locomotory behavior(GO:0007626) |
| 0.1 | 1.9 | GO:0048512 | circadian behavior(GO:0048512) |
| 0.1 | 1.2 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.1 | 1.3 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 2.1 | GO:0002718 | regulation of cytokine production involved in immune response(GO:0002718) |
| 0.1 | 5.8 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 1.1 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 3.2 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.2 | GO:0061311 | cell surface receptor signaling pathway involved in heart development(GO:0061311) |
| 0.1 | 0.2 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 0.1 | 0.2 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.1 | 0.4 | GO:0002155 | thyroid hormone mediated signaling pathway(GO:0002154) regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.1 | 2.0 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.1 | 0.8 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 1.3 | GO:0072171 | branching involved in ureteric bud morphogenesis(GO:0001658) ureteric bud morphogenesis(GO:0060675) mesonephric tubule morphogenesis(GO:0072171) |
| 0.1 | 1.8 | GO:0003197 | endocardial cushion development(GO:0003197) |
| 0.1 | 3.9 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 1.2 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.1 | 3.4 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.1 | 2.0 | GO:0003091 | renal water homeostasis(GO:0003091) |
| 0.1 | 0.9 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 0.5 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.1 | 3.3 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.1 | 0.4 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:0060003 | copper ion export(GO:0060003) |
| 0.0 | 1.6 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 6.0 | GO:0042594 | response to starvation(GO:0042594) |
| 0.0 | 0.5 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.2 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 1.0 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 1.3 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 1.1 | GO:0033273 | response to vitamin(GO:0033273) |
| 0.0 | 0.8 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.3 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 2.8 | GO:0075733 | intracellular transport of virus(GO:0075733) multi-organism intracellular transport(GO:1902583) |
| 0.0 | 0.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.6 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.2 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.0 | 1.5 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.1 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.0 | 0.4 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 1.0 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 3.9 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.3 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 2.0 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.3 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.9 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.3 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.2 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.7 | GO:0072384 | organelle transport along microtubule(GO:0072384) |
| 0.0 | 1.7 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 1.5 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.2 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.4 | GO:0006040 | amino sugar metabolic process(GO:0006040) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 17.9 | GO:0044609 | DBIRD complex(GO:0044609) |
| 4.6 | 13.9 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 4.0 | 52.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 3.6 | 14.4 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 3.2 | 12.8 | GO:0048179 | activin receptor complex(GO:0048179) |
| 2.9 | 23.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 2.1 | 10.4 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 2.0 | 6.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 1.8 | 5.5 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 1.8 | 10.6 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 1.7 | 3.4 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 1.6 | 4.9 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 1.6 | 6.2 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 1.5 | 19.7 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 1.4 | 4.3 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 1.3 | 3.8 | GO:0032116 | SMC loading complex(GO:0032116) |
| 1.2 | 9.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 1.1 | 6.8 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 1.1 | 8.6 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 1.0 | 12.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 1.0 | 5.8 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 1.0 | 19.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.9 | 3.8 | GO:0070695 | FHF complex(GO:0070695) |
| 0.9 | 10.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.9 | 5.5 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.9 | 4.4 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.9 | 3.5 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.8 | 5.9 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.8 | 3.4 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.8 | 13.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.8 | 2.4 | GO:0031251 | PAN complex(GO:0031251) |
| 0.8 | 11.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.7 | 3.7 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.7 | 4.0 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.7 | 7.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.6 | 7.8 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.6 | 44.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.6 | 3.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.6 | 8.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.6 | 10.9 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.6 | 21.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.6 | 3.4 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.6 | 17.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.6 | 29.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.6 | 3.9 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.5 | 2.7 | GO:0044308 | axonal spine(GO:0044308) |
| 0.5 | 3.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.5 | 10.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.5 | 12.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.5 | 4.3 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.5 | 2.8 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.4 | 5.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.4 | 43.8 | GO:0005901 | caveola(GO:0005901) |
| 0.4 | 2.5 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.4 | 3.7 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.4 | 7.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.4 | 5.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.4 | 2.7 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.4 | 7.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.4 | 20.4 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.3 | 1.4 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.3 | 2.4 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.3 | 2.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.3 | 9.7 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.3 | 2.5 | GO:0034464 | BBSome(GO:0034464) |
| 0.3 | 2.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.3 | 2.4 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.3 | 4.8 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.3 | 3.9 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.3 | 5.9 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.3 | 34.1 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.3 | 26.1 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.3 | 1.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.3 | 4.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.3 | 6.1 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.3 | 2.9 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.3 | 2.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.3 | 6.8 | GO:0044853 | plasma membrane raft(GO:0044853) |
| 0.2 | 5.9 | GO:0032982 | myosin filament(GO:0032982) |
| 0.2 | 2.0 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.2 | 68.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.2 | 1.6 | GO:0060091 | kinocilium(GO:0060091) |
| 0.2 | 8.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.2 | 3.0 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.2 | 17.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.2 | 5.5 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.2 | 8.5 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 16.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.2 | 8.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.2 | 4.6 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.2 | 1.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.2 | 14.5 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.2 | 6.6 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.2 | 2.7 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.2 | 11.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.2 | 3.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 0.8 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.2 | 12.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.2 | 7.2 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.2 | 0.5 | GO:0044292 | dendrite terminus(GO:0044292) dendritic growth cone(GO:0044294) |
| 0.2 | 1.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 1.5 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 3.0 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 3.1 | GO:0031094 | platelet dense tubular network(GO:0031094) platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 2.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 1.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 21.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 2.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 12.8 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.1 | 14.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 9.8 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 2.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 25.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 0.9 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 3.1 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 6.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 7.2 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.1 | 2.6 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 2.4 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.1 | 1.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 1.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 16.5 | GO:0031674 | I band(GO:0031674) |
| 0.1 | 9.8 | GO:0044215 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.1 | 186.4 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.1 | 2.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 2.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 1.3 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 0.8 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 3.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 0.5 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.1 | 1.6 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 1.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.6 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.1 | 5.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.2 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.1 | 5.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 2.2 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 3.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 1.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 5.6 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 45.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 2.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.4 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 7.9 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 2.2 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.9 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.8 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.3 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.4 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.4 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.2 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 15.6 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 4.4 | 17.4 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 4.1 | 20.3 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 3.7 | 11.2 | GO:0031696 | alpha-2C adrenergic receptor binding(GO:0031696) |
| 3.7 | 33.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 3.6 | 14.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 3.4 | 3.4 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 3.2 | 9.7 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 2.9 | 8.8 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 2.9 | 17.5 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 2.9 | 8.7 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 2.9 | 5.7 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 2.5 | 7.5 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 2.1 | 12.8 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 2.1 | 23.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 2.1 | 14.8 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 2.1 | 12.7 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 2.0 | 15.8 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 1.9 | 5.7 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) |
| 1.9 | 24.7 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 1.8 | 5.3 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 1.7 | 6.8 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 1.6 | 6.6 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 1.6 | 4.8 | GO:0015203 | polyamine transmembrane transporter activity(GO:0015203) putrescine transmembrane transporter activity(GO:0015489) |
| 1.5 | 10.7 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 1.5 | 6.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 1.5 | 4.4 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 1.4 | 21.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 1.4 | 4.3 | GO:0008988 | rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 1.4 | 10.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 1.4 | 4.3 | GO:0070039 | rRNA (guanosine-2'-O-)-methyltransferase activity(GO:0070039) |
| 1.4 | 7.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 1.4 | 4.2 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 1.4 | 17.7 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 1.3 | 10.3 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 1.3 | 3.8 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 1.2 | 3.6 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 1.2 | 25.0 | GO:0005112 | Notch binding(GO:0005112) |
| 1.2 | 11.8 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 1.2 | 3.5 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 1.2 | 1.2 | GO:0035276 | ethanol binding(GO:0035276) |
| 1.1 | 7.9 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 1.1 | 3.4 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 1.1 | 5.4 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 1.1 | 4.3 | GO:0004979 | beta-endorphin receptor activity(GO:0004979) morphine receptor activity(GO:0038047) |
| 1.1 | 5.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 1.1 | 3.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 1.1 | 3.2 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 1.0 | 9.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 1.0 | 4.2 | GO:0004773 | steryl-sulfatase activity(GO:0004773) |
| 1.0 | 4.1 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 1.0 | 7.2 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 1.0 | 31.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 1.0 | 16.8 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 1.0 | 2.9 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 1.0 | 2.9 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 1.0 | 14.3 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.9 | 14.0 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.9 | 73.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.9 | 5.5 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.9 | 12.9 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.9 | 6.2 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.9 | 10.6 | GO:0048185 | activin binding(GO:0048185) |
| 0.9 | 2.6 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.9 | 5.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.8 | 6.8 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.8 | 4.2 | GO:1901474 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) azole transmembrane transporter activity(GO:1901474) |
| 0.8 | 10.9 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.8 | 3.4 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.8 | 4.9 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.8 | 3.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.8 | 2.4 | GO:0004960 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.8 | 16.0 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.8 | 4.0 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.8 | 2.3 | GO:0015181 | L-ornithine transmembrane transporter activity(GO:0000064) arginine transmembrane transporter activity(GO:0015181) |
| 0.8 | 3.8 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.7 | 3.7 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.7 | 0.7 | GO:0045118 | azole transporter activity(GO:0045118) |
| 0.7 | 17.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.7 | 9.9 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.7 | 6.2 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.7 | 12.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.7 | 3.4 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.7 | 2.0 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.7 | 4.0 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.7 | 4.7 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.7 | 2.6 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.6 | 1.9 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.6 | 4.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.6 | 7.6 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.6 | 20.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.6 | 2.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.6 | 7.7 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.6 | 1.8 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.6 | 1.7 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.6 | 2.9 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.6 | 12.1 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.6 | 5.7 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.6 | 12.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.6 | 14.6 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.6 | 2.8 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.6 | 4.5 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.5 | 4.9 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.5 | 3.3 | GO:0099529 | postsynaptic neurotransmitter receptor activity(GO:0098960) neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.5 | 3.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.5 | 3.7 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.5 | 4.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.5 | 7.9 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.5 | 7.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.5 | 2.6 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.5 | 1.6 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.5 | 2.6 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.5 | 2.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.5 | 2.5 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.5 | 3.5 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.5 | 6.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.5 | 8.0 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.5 | 2.5 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.5 | 8.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.5 | 1.9 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.5 | 6.3 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.5 | 4.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.5 | 2.4 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.5 | 6.7 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.4 | 3.1 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.4 | 4.5 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.4 | 1.3 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.4 | 3.6 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.4 | 5.8 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.4 | 4.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.4 | 6.6 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.4 | 12.3 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.4 | 1.7 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.4 | 3.9 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.4 | 12.4 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.4 | 3.0 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.4 | 1.3 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.4 | 4.9 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.4 | 5.6 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.4 | 6.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.4 | 2.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.4 | 12.2 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.4 | 1.2 | GO:0051379 | beta-adrenergic receptor activity(GO:0004939) epinephrine binding(GO:0051379) norepinephrine binding(GO:0051380) |
| 0.4 | 6.0 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.4 | 9.8 | GO:0001848 | complement binding(GO:0001848) |
| 0.4 | 24.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.4 | 7.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.4 | 1.1 | GO:0046921 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.4 | 3.0 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.4 | 3.7 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.4 | 20.9 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.4 | 4.3 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.3 | 1.7 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.3 | 3.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.3 | 3.0 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.3 | 1.3 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.3 | 1.0 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.3 | 4.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.3 | 2.8 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.3 | 0.9 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.3 | 1.8 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.3 | 11.3 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.3 | 3.7 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.3 | 8.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.3 | 0.8 | GO:0031753 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.3 | 1.1 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.3 | 76.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.3 | 2.5 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.3 | 1.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.3 | 3.0 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.3 | 4.4 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.3 | 1.9 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.3 | 5.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.3 | 2.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.3 | 3.7 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.3 | 3.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.3 | 0.3 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.3 | 2.3 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.3 | 1.3 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.2 | 3.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.2 | 7.7 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.2 | 8.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 4.1 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.2 | 0.9 | GO:0043273 | CTPase activity(GO:0043273) |
| 0.2 | 3.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 0.9 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 7.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 1.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.2 | 0.6 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.2 | 3.4 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 2.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.2 | 1.2 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.2 | 3.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 1.9 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 6.0 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.2 | 19.1 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.2 | 5.0 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.2 | 5.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 4.6 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.2 | 2.0 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.2 | 5.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.2 | 1.3 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.2 | 7.0 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.2 | 0.9 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.2 | 18.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 5.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.2 | 2.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 2.2 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.2 | 2.0 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.2 | 2.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 2.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 3.0 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.2 | 3.3 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.2 | 1.6 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.2 | 12.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 2.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 2.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 0.5 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 1.0 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.2 | 5.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 3.0 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.2 | 4.2 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.2 | 3.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 0.5 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.2 | 1.9 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 62.8 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.2 | 1.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 2.7 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 11.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.6 | GO:0015375 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 1.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 2.6 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 2.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.9 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 0.7 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 4.6 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 1.2 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.1 | 2.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.4 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 16.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 9.1 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 3.7 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 3.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 8.3 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 2.1 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 1.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 2.7 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 3.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 1.0 | GO:0042379 | chemokine receptor binding(GO:0042379) |
| 0.1 | 6.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.2 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.1 | 0.8 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.1 | 1.9 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 3.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 6.0 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 1.6 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 2.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.9 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 1.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 49.9 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.1 | 5.5 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 6.7 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 0.2 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 2.2 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.1 | 0.7 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 4.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 1.6 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 1.0 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.1 | 6.0 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.1 | 0.9 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 1.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 4.1 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 2.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 2.6 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 0.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 2.6 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 5.9 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 0.4 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 0.4 | GO:0005497 | androgen binding(GO:0005497) |
| 0.1 | 9.7 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 0.9 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.1 | 1.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 3.2 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 0.5 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 1.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.2 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 1.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 1.0 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 1.5 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 0.9 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.1 | 1.0 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.6 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 3.5 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.2 | GO:0048403 | neurotrophin receptor activity(GO:0005030) brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 1.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.5 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 2.4 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.6 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 21.2 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.1 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.0 | 0.8 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 1.2 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 2.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.3 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.7 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.4 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 2.7 | GO:0005506 | iron ion binding(GO:0005506) |
| 0.0 | 2.3 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 1.7 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.1 | GO:0008948 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) oxaloacetate decarboxylase activity(GO:0008948) |
| 0.0 | 2.1 | GO:0030246 | carbohydrate binding(GO:0030246) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 78.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 1.0 | 51.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.9 | 44.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.9 | 4.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.8 | 17.0 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.7 | 7.4 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.7 | 7.4 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.7 | 12.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.7 | 21.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.6 | 33.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.5 | 4.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.5 | 7.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.4 | 93.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.4 | 30.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.4 | 10.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.4 | 5.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.4 | 25.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.4 | 15.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.3 | 3.4 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.3 | 2.9 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.3 | 1.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.3 | 6.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.3 | 6.8 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.3 | 3.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.3 | 8.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.3 | 13.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.3 | 11.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.2 | 42.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 3.9 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 59.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 14.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 6.6 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.2 | 4.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 4.9 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.2 | 2.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 1.8 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 5.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.2 | 3.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.2 | 64.1 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.2 | 10.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.2 | 6.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.2 | 7.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.2 | 10.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 10.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 4.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 4.9 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 4.5 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 5.0 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 4.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 0.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 1.8 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 0.4 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 1.8 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 2.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 4.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 2.8 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.1 | 3.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 3.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 1.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.0 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.6 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.3 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 52.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 1.3 | 23.9 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 1.1 | 14.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.9 | 67.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.8 | 4.2 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.8 | 2.3 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.7 | 7.3 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.7 | 12.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.7 | 11.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.7 | 16.9 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.7 | 11.0 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.6 | 15.0 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.6 | 21.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.6 | 12.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.5 | 7.6 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.5 | 4.2 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.5 | 2.6 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.5 | 31.9 | REACTOME GAB1 SIGNALOSOME | Genes involved in GAB1 signalosome |
| 0.4 | 3.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.4 | 7.0 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.4 | 7.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.4 | 9.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.4 | 7.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.4 | 4.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.4 | 9.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.4 | 10.9 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.3 | 11.2 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.3 | 3.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.3 | 10.1 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.3 | 5.0 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.3 | 3.7 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.3 | 12.8 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.3 | 14.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.3 | 6.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.3 | 4.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.3 | 8.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.3 | 30.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.3 | 9.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.3 | 11.1 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.3 | 0.8 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.3 | 11.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.3 | 9.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.3 | 9.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.3 | 3.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 13.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.3 | 5.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 2.5 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.2 | 14.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.2 | 15.3 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.2 | 14.3 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.2 | 4.8 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.2 | 2.8 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.2 | 3.7 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.2 | 2.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.2 | 5.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 5.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.2 | 3.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 3.0 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 1.6 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.2 | 5.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 4.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 3.9 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 5.0 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.2 | 0.8 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.2 | 2.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.2 | 1.8 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 2.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 4.3 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.1 | 5.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 6.6 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |
| 0.1 | 19.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 5.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 4.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 3.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 0.9 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 2.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 1.0 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 22.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 2.6 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 0.6 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 2.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 10.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 10.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 0.8 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 2.0 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.1 | 2.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.5 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 1.9 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 1.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 2.2 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 3.5 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 6.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 3.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 1.6 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 4.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 0.9 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 2.6 | REACTOME RNA POL III TRANSCRIPTION | Genes involved in RNA Polymerase III Transcription |
| 0.1 | 4.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 0.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 2.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 1.1 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 3.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.9 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 1.1 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 2.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.8 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 5.8 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 2.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.3 | REACTOME BIOLOGICAL OXIDATIONS | Genes involved in Biological oxidations |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |