Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for ZNF274
Z-value: 4.78
Transcription factors associated with ZNF274
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF274
|
ENSG00000171606.18 | ZNF274 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF274 | hg38_v1_chr19_+_58183375_58183494 | 0.45 | 3.0e-12 | Click! |
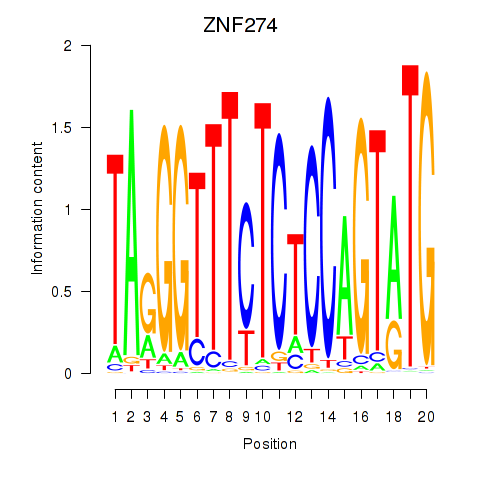
Activity profile of ZNF274 motif
Sorted Z-values of ZNF274 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF274
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.8 | 43.0 | GO:0002188 | translation reinitiation(GO:0002188) |
| 2.1 | 8.5 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 1.4 | 12.8 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 1.2 | 7.0 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.9 | 4.6 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.9 | 4.3 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.7 | 6.9 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.6 | 19.7 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.5 | 17.4 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) |
| 0.5 | 7.1 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.4 | 2.2 | GO:1902523 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.4 | 35.4 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.2 | 15.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 10.0 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.1 | 2.1 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.1 | 4.6 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 7.3 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 14.8 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 5.2 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 0.1 | GO:0031438 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 4.0 | GO:0007565 | female pregnancy(GO:0007565) |
| 0.0 | 0.5 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 15.3 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 1.1 | 14.8 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.6 | 43.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.5 | 10.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.4 | 12.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.2 | 6.9 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.2 | 4.3 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.1 | 19.7 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 9.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 5.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 4.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 2.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 12.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.5 | GO:0051233 | spindle midzone(GO:0051233) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 9.9 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 2.5 | 7.4 | GO:0048030 | disaccharide binding(GO:0048030) |
| 1.6 | 12.8 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 1.4 | 7.0 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 1.2 | 8.5 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.9 | 4.6 | GO:0004471 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) oxaloacetate decarboxylase activity(GO:0008948) |
| 0.8 | 19.7 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.8 | 6.9 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.7 | 15.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.6 | 43.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.4 | 4.6 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.4 | 10.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.3 | 17.4 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.3 | 35.4 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.2 | 14.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.2 | 4.3 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 2.1 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 5.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 2.2 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 27.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.4 | 19.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.3 | 8.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 2.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 5.2 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 7.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 4.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 12.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.3 | 19.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.2 | 15.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 5.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 4.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 6.9 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 10.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 14.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 2.1 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 4.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 13.6 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 8.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |