Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for ZNF333
Z-value: 2.73
Transcription factors associated with ZNF333
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF333
|
ENSG00000160961.12 | ZNF333 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF333 | hg38_v1_chr19_+_14690037_14690105 | 0.13 | 5.6e-02 | Click! |
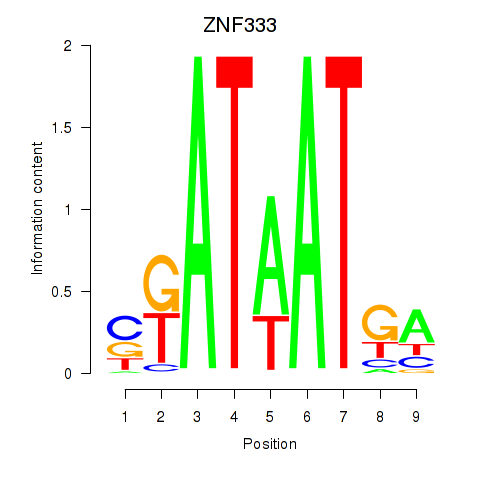
Activity profile of ZNF333 motif
Sorted Z-values of ZNF333 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF333
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_200440649 | 25.30 |
ENST00000366118.2
|
SPATS2L
|
spermatogenesis associated serine rich 2 like |
| chr6_+_29301701 | 8.22 |
ENST00000641895.1
|
OR14J1
|
olfactory receptor family 14 subfamily J member 1 |
| chr4_-_46909235 | 7.51 |
ENST00000505102.1
ENST00000355591.8 |
COX7B2
|
cytochrome c oxidase subunit 7B2 |
| chr4_-_46909206 | 7.32 |
ENST00000396533.5
|
COX7B2
|
cytochrome c oxidase subunit 7B2 |
| chr18_+_74148508 | 6.61 |
ENST00000580087.5
ENST00000169551.11 |
TIMM21
|
translocase of inner mitochondrial membrane 21 |
| chr19_+_44905785 | 6.33 |
ENST00000446996.5
ENST00000252486.9 ENST00000434152.5 |
APOE
|
apolipoprotein E |
| chr7_-_116030735 | 5.77 |
ENST00000393485.5
|
TFEC
|
transcription factor EC |
| chr5_+_157743703 | 5.31 |
ENST00000286307.6
|
LSM11
|
LSM11, U7 small nuclear RNA associated |
| chr1_+_19312341 | 5.07 |
ENST00000400548.6
|
SLC66A1
|
solute carrier family 66 member 1 |
| chr6_+_131573219 | 5.00 |
ENST00000356962.2
ENST00000368087.8 ENST00000673427.1 ENST00000640973.1 |
ARG1
|
arginase 1 |
| chr11_-_5249836 | 4.92 |
ENST00000632727.1
ENST00000330597.5 |
HBG1
|
hemoglobin subunit gamma 1 |
| chr3_-_42875871 | 4.87 |
ENST00000316161.6
ENST00000437102.1 |
CYP8B1
|
cytochrome P450 family 8 subfamily B member 1 |
| chr7_-_116030750 | 4.85 |
ENST00000265440.12
ENST00000320239.11 |
TFEC
|
transcription factor EC |
| chr16_+_84294823 | 4.55 |
ENST00000568638.1
|
WFDC1
|
WAP four-disulfide core domain 1 |
| chr1_+_248231417 | 4.46 |
ENST00000641868.1
|
OR2M4
|
olfactory receptor family 2 subfamily M member 4 |
| chr1_+_19312296 | 4.44 |
ENST00000375155.7
ENST00000375153.8 |
SLC66A1
|
solute carrier family 66 member 1 |
| chr12_-_70637405 | 4.30 |
ENST00000548122.2
ENST00000551525.5 ENST00000550358.5 ENST00000334414.11 |
PTPRB
|
protein tyrosine phosphatase receptor type B |
| chr16_+_84294853 | 4.25 |
ENST00000219454.10
|
WFDC1
|
WAP four-disulfide core domain 1 |
| chr5_+_137889469 | 4.21 |
ENST00000290431.5
|
PKD2L2
|
polycystin 2 like 2, transient receptor potential cation channel |
| chr3_+_35679614 | 4.11 |
ENST00000474696.5
ENST00000412048.5 ENST00000396482.6 ENST00000432682.5 |
ARPP21
|
cAMP regulated phosphoprotein 21 |
| chr7_-_83649097 | 4.03 |
ENST00000643230.2
|
SEMA3E
|
semaphorin 3E |
| chr2_-_127220293 | 3.91 |
ENST00000664447.2
ENST00000409327.2 |
CYP27C1
|
cytochrome P450 family 27 subfamily C member 1 |
| chr9_-_94593810 | 3.84 |
ENST00000375337.4
|
FBP2
|
fructose-bisphosphatase 2 |
| chr5_+_73626158 | 3.77 |
ENST00000296794.10
ENST00000545377.5 ENST00000509848.5 ENST00000513042.7 |
ARHGEF28
|
Rho guanine nucleotide exchange factor 28 |
| chr12_+_112125531 | 3.71 |
ENST00000549358.5
ENST00000257604.9 ENST00000548092.5 ENST00000412615.7 ENST00000552896.1 |
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chrX_-_19970298 | 3.67 |
ENST00000379687.7
ENST00000379682.8 |
BCLAF3
|
BCLAF1 and THRAP3 family member 3 |
| chr9_-_107489754 | 3.66 |
ENST00000610832.1
ENST00000374672.5 |
KLF4
|
Kruppel like factor 4 |
| chr14_+_20110739 | 3.64 |
ENST00000641386.2
ENST00000641633.2 |
OR4K17
|
olfactory receptor family 4 subfamily K member 17 |
| chr7_+_29122274 | 3.33 |
ENST00000582692.2
ENST00000644824.1 |
ENSG00000285412.1
ENSG00000285162.1
|
novel transcript, antisense to CPVL chimerin 2 |
| chr19_-_44500503 | 3.21 |
ENST00000587047.1
ENST00000391956.8 ENST00000221327.8 ENST00000592529.6 ENST00000591064.1 |
ZNF180
|
zinc finger protein 180 |
| chrX_-_30577759 | 3.10 |
ENST00000378962.4
|
TASL
|
TLR adaptor interacting with endolysosomal SLC15A4 |
| chr3_+_196867856 | 3.05 |
ENST00000445299.6
ENST00000323460.10 ENST00000419026.5 |
SENP5
|
SUMO specific peptidase 5 |
| chr20_+_59940362 | 3.03 |
ENST00000360816.8
|
FAM217B
|
family with sequence similarity 217 member B |
| chr5_+_141421064 | 2.96 |
ENST00000518882.2
|
PCDHGA11
|
protocadherin gamma subfamily A, 11 |
| chr3_+_48241046 | 2.91 |
ENST00000427617.6
ENST00000412564.5 ENST00000354698.8 ENST00000440261.6 |
ZNF589
|
zinc finger protein 589 |
| chr2_+_79185231 | 2.85 |
ENST00000466387.5
|
CTNNA2
|
catenin alpha 2 |
| chr20_-_45972171 | 2.81 |
ENST00000322927.3
|
ZNF335
|
zinc finger protein 335 |
| chr9_-_111036207 | 2.76 |
ENST00000541779.5
|
LPAR1
|
lysophosphatidic acid receptor 1 |
| chr8_+_18391276 | 2.76 |
ENST00000286479.4
ENST00000520116.1 |
NAT2
|
N-acetyltransferase 2 |
| chr5_+_137889437 | 2.74 |
ENST00000508638.5
ENST00000508883.6 ENST00000502810.5 |
PKD2L2
|
polycystin 2 like 2, transient receptor potential cation channel |
| chr9_-_19786928 | 2.71 |
ENST00000341998.6
ENST00000286344.3 |
SLC24A2
|
solute carrier family 24 member 2 |
| chr3_+_38282294 | 2.66 |
ENST00000466887.5
ENST00000448498.6 |
SLC22A14
|
solute carrier family 22 member 14 |
| chr1_+_196888014 | 2.66 |
ENST00000367416.6
ENST00000608469.6 ENST00000251424.8 ENST00000367418.2 |
CFHR4
|
complement factor H related 4 |
| chr2_-_177888728 | 2.57 |
ENST00000389683.7
|
PDE11A
|
phosphodiesterase 11A |
| chr6_+_50818701 | 2.55 |
ENST00000344788.7
|
TFAP2B
|
transcription factor AP-2 beta |
| chr5_+_140827950 | 2.54 |
ENST00000378126.4
ENST00000529310.6 ENST00000527624.1 |
PCDHA6
|
protocadherin alpha 6 |
| chrX_+_11293411 | 2.43 |
ENST00000348912.4
ENST00000380714.7 ENST00000380712.7 |
AMELX
|
amelogenin X-linked |
| chrX_-_21758021 | 2.40 |
ENST00000646008.1
|
SMPX
|
small muscle protein X-linked |
| chrX_-_21758097 | 2.40 |
ENST00000379494.4
|
SMPX
|
small muscle protein X-linked |
| chr11_-_14971179 | 2.34 |
ENST00000486207.5
|
CALCA
|
calcitonin related polypeptide alpha |
| chr10_-_67665642 | 2.30 |
ENST00000682945.1
ENST00000330298.6 ENST00000682758.1 |
CTNNA3
|
catenin alpha 3 |
| chr8_-_30912998 | 2.27 |
ENST00000643185.2
|
TEX15
|
testis expressed 15, meiosis and synapsis associated |
| chr6_-_32941018 | 2.14 |
ENST00000418107.3
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr5_-_78985249 | 2.10 |
ENST00000565165.2
|
ARSB
|
arylsulfatase B |
| chr6_+_29585121 | 2.02 |
ENST00000641840.1
|
OR2H2
|
olfactory receptor family 2 subfamily H member 2 |
| chr17_-_3433841 | 2.01 |
ENST00000248384.1
|
OR1E2
|
olfactory receptor family 1 subfamily E member 2 |
| chr9_+_32551670 | 2.00 |
ENST00000450093.3
|
SMIM27
|
small integral membrane protein 27 |
| chr2_+_29113989 | 1.78 |
ENST00000404424.5
|
CLIP4
|
CAP-Gly domain containing linker protein family member 4 |
| chr14_+_20768393 | 1.75 |
ENST00000326783.4
|
EDDM3B
|
epididymal protein 3B |
| chr5_+_141421020 | 1.75 |
ENST00000622044.1
ENST00000398587.7 |
PCDHGA11
|
protocadherin gamma subfamily A, 11 |
| chr11_+_18412292 | 1.75 |
ENST00000541669.6
ENST00000280704.8 |
LDHC
|
lactate dehydrogenase C |
| chr6_+_50818857 | 1.70 |
ENST00000393655.4
|
TFAP2B
|
transcription factor AP-2 beta |
| chr7_+_144069811 | 1.64 |
ENST00000641663.1
|
OR2A25
|
olfactory receptor family 2 subfamily A member 25 |
| chr14_+_85530163 | 1.63 |
ENST00000554746.1
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr2_+_232662733 | 1.59 |
ENST00000410095.5
ENST00000611312.1 |
EFHD1
|
EF-hand domain family member D1 |
| chr14_+_85530127 | 1.58 |
ENST00000330753.6
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr7_-_80512041 | 1.56 |
ENST00000398291.4
|
GNAT3
|
G protein subunit alpha transducin 3 |
| chr14_+_23372809 | 1.50 |
ENST00000397242.2
ENST00000329715.2 |
IL25
|
interleukin 25 |
| chr2_-_2331336 | 1.49 |
ENST00000648933.1
ENST00000644820.1 |
MYT1L
|
myelin transcription factor 1 like |
| chr1_+_213987929 | 1.49 |
ENST00000498508.6
ENST00000366958.9 |
PROX1
|
prospero homeobox 1 |
| chr12_-_48999363 | 1.36 |
ENST00000421952.3
|
DDN
|
dendrin |
| chr17_-_3127551 | 1.31 |
ENST00000328890.3
|
OR1G1
|
olfactory receptor family 1 subfamily G member 1 |
| chr6_+_131135408 | 1.31 |
ENST00000431975.7
ENST00000683794.1 |
AKAP7
|
A-kinase anchoring protein 7 |
| chr4_+_70242583 | 1.30 |
ENST00000304954.3
|
CSN3
|
casein kappa |
| chr11_+_33039996 | 1.24 |
ENST00000432887.5
ENST00000528898.1 ENST00000531632.6 |
TCP11L1
|
t-complex 11 like 1 |
| chrX_+_100584928 | 1.23 |
ENST00000373031.5
|
TNMD
|
tenomodulin |
| chr3_-_114199407 | 1.20 |
ENST00000460779.5
|
DRD3
|
dopamine receptor D3 |
| chr4_+_105552611 | 1.20 |
ENST00000265154.6
ENST00000420470.3 |
ARHGEF38
|
Rho guanine nucleotide exchange factor 38 |
| chr16_-_20691256 | 1.09 |
ENST00000307493.8
|
ACSM1
|
acyl-CoA synthetase medium chain family member 1 |
| chrX_-_70260199 | 1.02 |
ENST00000374519.4
|
P2RY4
|
pyrimidinergic receptor P2Y4 |
| chr5_-_16508951 | 1.00 |
ENST00000682628.1
|
RETREG1
|
reticulophagy regulator 1 |
| chr1_-_169630115 | 1.00 |
ENST00000263686.11
ENST00000367788.6 |
SELP
|
selectin P |
| chr5_-_16508858 | 0.98 |
ENST00000684456.1
|
RETREG1
|
reticulophagy regulator 1 |
| chr5_-_16508812 | 0.96 |
ENST00000683414.1
|
RETREG1
|
reticulophagy regulator 1 |
| chr20_+_45207025 | 0.95 |
ENST00000372781.4
|
SEMG1
|
semenogelin 1 |
| chr1_+_54548217 | 0.94 |
ENST00000343744.7
ENST00000371316.3 |
ACOT11
|
acyl-CoA thioesterase 11 |
| chr4_+_69931066 | 0.93 |
ENST00000246891.9
|
CSN1S1
|
casein alpha s1 |
| chrX_-_49264668 | 0.93 |
ENST00000455775.7
ENST00000652559.1 ENST00000376207.10 ENST00000557224.6 ENST00000684155.1 ENST00000376199.7 |
FOXP3
|
forkhead box P3 |
| chr4_+_109827963 | 0.82 |
ENST00000317735.7
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr12_-_7695752 | 0.80 |
ENST00000329913.4
|
GDF3
|
growth differentiation factor 3 |
| chr2_+_102311502 | 0.69 |
ENST00000404917.6
ENST00000410040.5 |
IL1RL1
IL18R1
|
interleukin 1 receptor like 1 interleukin 18 receptor 1 |
| chr18_+_48539017 | 0.67 |
ENST00000256413.8
|
CTIF
|
cap binding complex dependent translation initiation factor |
| chr5_-_27038576 | 0.65 |
ENST00000511822.1
ENST00000231021.9 |
CDH9
|
cadherin 9 |
| chr2_-_144517663 | 0.60 |
ENST00000427902.5
ENST00000462355.2 ENST00000470879.5 ENST00000409487.7 ENST00000435831.5 ENST00000630572.2 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr5_-_147782518 | 0.51 |
ENST00000507386.5
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2 |
| chr3_-_116444983 | 0.51 |
ENST00000333617.8
|
LSAMP
|
limbic system associated membrane protein |
| chr2_+_216498831 | 0.45 |
ENST00000491306.6
ENST00000600880.5 ENST00000446558.5 |
RPL37A
|
ribosomal protein L37a |
| chr1_-_27374816 | 0.44 |
ENST00000270879.9
ENST00000354982.2 |
FCN3
|
ficolin 3 |
| chr2_+_88885397 | 0.41 |
ENST00000390243.2
|
IGKV4-1
|
immunoglobulin kappa variable 4-1 |
| chr12_-_10807286 | 0.38 |
ENST00000240615.3
|
TAS2R8
|
taste 2 receptor member 8 |
| chr1_+_59297057 | 0.38 |
ENST00000303721.12
|
FGGY
|
FGGY carbohydrate kinase domain containing |
| chr1_+_159439722 | 0.36 |
ENST00000641630.1
ENST00000423932.6 |
OR10J1
|
olfactory receptor family 10 subfamily J member 1 |
| chr1_-_51297990 | 0.24 |
ENST00000530004.5
|
TTC39A
|
tetratricopeptide repeat domain 39A |
| chr11_-_72793592 | 0.24 |
ENST00000536377.5
ENST00000359373.9 |
STARD10
ARAP1
|
StAR related lipid transfer domain containing 10 ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr14_+_23630109 | 0.23 |
ENST00000432832.6
|
DHRS2
|
dehydrogenase/reductase 2 |
| chr10_-_121596117 | 0.14 |
ENST00000351936.11
|
FGFR2
|
fibroblast growth factor receptor 2 |
| chr12_+_59664677 | 0.14 |
ENST00000548610.5
|
SLC16A7
|
solute carrier family 16 member 7 |
| chr8_+_127409026 | 0.14 |
ENST00000465342.4
|
POU5F1B
|
POU class 5 homeobox 1B |
| chr7_-_25228485 | 0.12 |
ENST00000222674.2
|
NPVF
|
neuropeptide VF precursor |
| chr13_-_71867192 | 0.11 |
ENST00000611519.4
ENST00000620444.4 ENST00000613252.5 |
DACH1
|
dachshund family transcription factor 1 |
| chr5_-_78985288 | 0.09 |
ENST00000264914.10
|
ARSB
|
arylsulfatase B |
| chr1_-_197067234 | 0.08 |
ENST00000367412.2
|
F13B
|
coagulation factor XIII B chain |
| chr8_+_40153475 | 0.07 |
ENST00000315792.5
|
TCIM
|
transcriptional and immune response regulator |
| chr17_-_35119733 | 0.06 |
ENST00000460118.6
ENST00000335858.11 |
RAD51D
|
RAD51 paralog D |
| chr2_-_213151590 | 0.04 |
ENST00000374319.8
ENST00000457361.5 ENST00000451136.6 ENST00000434687.6 |
IKZF2
|
IKAROS family zinc finger 2 |
| chrX_-_120311533 | 0.02 |
ENST00000440464.5
ENST00000519908.1 |
TMEM255A
|
transmembrane protein 255A |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 1.3 | 6.3 | GO:1902952 | positive regulation of postsynaptic membrane organization(GO:1901628) positive regulation of dendritic spine maintenance(GO:1902952) |
| 1.3 | 5.0 | GO:0043091 | L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 1.2 | 3.7 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 1.0 | 3.8 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.9 | 5.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.8 | 2.3 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.7 | 2.1 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.6 | 2.8 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.5 | 2.2 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.5 | 3.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.5 | 6.6 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.5 | 1.5 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.4 | 1.7 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 0.4 | 2.4 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.4 | 0.8 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.4 | 1.6 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.4 | 1.1 | GO:0018874 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 0.4 | 3.2 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.3 | 2.6 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.3 | 0.9 | GO:0032829 | tolerance induction dependent upon immune response(GO:0002461) regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) positive regulation of immature T cell proliferation(GO:0033091) |
| 0.3 | 1.2 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) gastric motility(GO:0035482) gastric emptying(GO:0035483) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.3 | 2.3 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.3 | 2.8 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.3 | 3.9 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.3 | 4.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.3 | 1.5 | GO:0009624 | response to nematode(GO:0009624) |
| 0.2 | 1.2 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.2 | 23.7 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.2 | 2.9 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.2 | 0.9 | GO:1902573 | positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.1 | 6.9 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.1 | 13.0 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 2.6 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 4.0 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.1 | 4.9 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.1 | 0.7 | GO:0071351 | negative regulation of T-helper 1 type immune response(GO:0002826) interleukin-18-mediated signaling pathway(GO:0035655) interleukin-33-mediated signaling pathway(GO:0038172) cellular response to interleukin-18(GO:0071351) |
| 0.1 | 1.0 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 2.3 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 1.0 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 8.8 | GO:0061045 | negative regulation of wound healing(GO:0061045) |
| 0.1 | 2.7 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 0.6 | GO:1902748 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 14.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 2.7 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.1 | 3.5 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.1 | 0.2 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) bile acid secretion(GO:0032782) |
| 0.1 | 2.8 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.1 | 0.4 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.8 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 7.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.5 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.4 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 5.0 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 3.2 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.1 | GO:0035602 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) |
| 0.0 | 0.5 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 2.6 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.9 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 1.3 | GO:0007595 | lactation(GO:0007595) |
| 0.0 | 0.1 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.1 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.3 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.9 | 14.8 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.8 | 4.8 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.8 | 5.3 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.5 | 6.6 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.3 | 4.9 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 2.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 3.7 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 1.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 7.2 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 1.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 1.0 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 2.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 4.3 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 2.8 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 1.6 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 2.9 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 2.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 2.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.8 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.8 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.7 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 5.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.3 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 1.3 | 3.9 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 1.0 | 3.8 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.9 | 2.8 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.8 | 8.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.8 | 2.4 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.7 | 3.7 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.7 | 2.2 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.6 | 2.3 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.5 | 3.1 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.5 | 14.0 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.5 | 2.7 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.3 | 5.0 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.3 | 1.0 | GO:0042806 | fucose binding(GO:0042806) |
| 0.3 | 4.9 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.3 | 2.8 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.3 | 2.3 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.3 | 4.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 1.7 | GO:0004459 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 0.7 | GO:0002113 | interleukin-33 binding(GO:0002113) interleukin-18 receptor activity(GO:0042008) |
| 0.2 | 2.6 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.2 | 1.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.2 | 15.4 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.2 | 3.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.2 | 1.4 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) |
| 0.1 | 1.5 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 2.7 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.1 | 1.0 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.1 | 4.9 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 0.4 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.1 | 1.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 4.2 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 3.2 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 5.3 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.1 | 0.9 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 8.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 2.1 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 0.8 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.9 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 6.9 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.2 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 1.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 5.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.6 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 10.6 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 2.7 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.0 | 5.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 2.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 4.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 10.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 6.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 2.8 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 4.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 2.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.4 | 5.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.3 | 6.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 21.6 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.2 | 3.7 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 2.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 4.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.8 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 2.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 3.8 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 2.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.0 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 1.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 2.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 2.1 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 2.8 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 1.1 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |