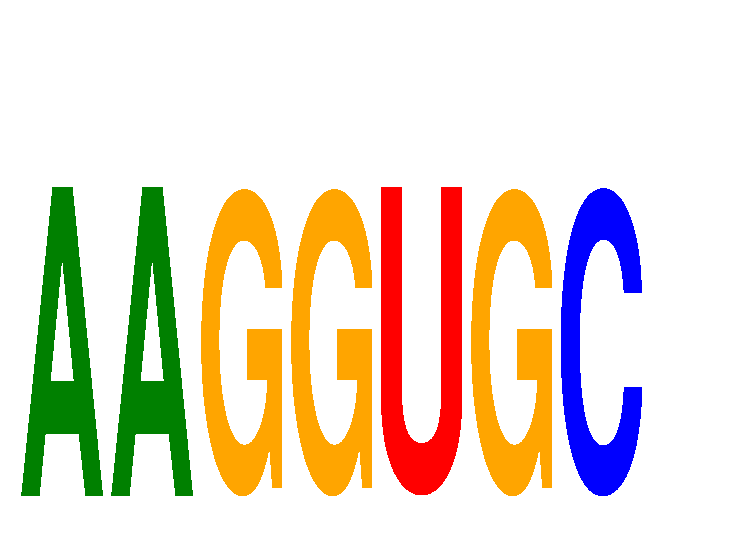Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for AAGGUGC
Z-value: 0.31
miRNA associated with seed AAGGUGC
| Name | miRBASE accession |
|---|---|
|
hsa-miR-18a-5p
|
MIMAT0000072 |
|
hsa-miR-18b-5p
|
MIMAT0001412 |
|
hsa-miR-4735-3p
|
MIMAT0019861 |
Activity profile of AAGGUGC motif
Sorted Z-values of AAGGUGC motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_39873268 | 0.76 |
ENST00000397591.2
ENST00000260356.5 |
THBS1
|
thrombospondin 1 |
| chr13_-_77601282 | 0.35 |
ENST00000355619.5
|
FBXL3
|
F-box and leucine-rich repeat protein 3 |
| chr12_+_4382917 | 0.32 |
ENST00000261254.3
|
CCND2
|
cyclin D2 |
| chr4_-_125633876 | 0.32 |
ENST00000504087.1
ENST00000515641.1 |
ANKRD50
|
ankyrin repeat domain 50 |
| chr3_-_113465065 | 0.32 |
ENST00000497255.1
ENST00000478020.1 ENST00000240922.3 ENST00000493900.1 |
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr2_-_9143786 | 0.32 |
ENST00000462696.1
ENST00000305997.3 |
MBOAT2
|
membrane bound O-acyltransferase domain containing 2 |
| chr12_+_69864129 | 0.31 |
ENST00000547219.1
ENST00000299293.2 ENST00000549921.1 ENST00000550316.1 ENST00000548154.1 ENST00000547414.1 ENST00000550389.1 ENST00000550937.1 ENST00000549092.1 ENST00000550169.1 |
FRS2
|
fibroblast growth factor receptor substrate 2 |
| chr1_+_173684047 | 0.31 |
ENST00000546011.1
ENST00000209884.4 |
KLHL20
|
kelch-like family member 20 |
| chr17_-_27621125 | 0.30 |
ENST00000579665.1
ENST00000225388.4 |
NUFIP2
|
nuclear fragile X mental retardation protein interacting protein 2 |
| chr3_-_123603137 | 0.29 |
ENST00000360304.3
ENST00000359169.1 ENST00000346322.5 ENST00000360772.3 |
MYLK
|
myosin light chain kinase |
| chr15_-_34628951 | 0.29 |
ENST00000397707.2
ENST00000560611.1 |
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr16_+_71879861 | 0.28 |
ENST00000427980.2
ENST00000568581.1 |
ATXN1L
IST1
|
ataxin 1-like increased sodium tolerance 1 homolog (yeast) |
| chr10_+_111767720 | 0.26 |
ENST00000356080.4
ENST00000277900.8 |
ADD3
|
adducin 3 (gamma) |
| chr9_-_99180597 | 0.26 |
ENST00000375256.4
|
ZNF367
|
zinc finger protein 367 |
| chr12_+_104458235 | 0.24 |
ENST00000229330.4
|
HCFC2
|
host cell factor C2 |
| chr2_-_26101374 | 0.22 |
ENST00000435504.4
|
ASXL2
|
additional sex combs like 2 (Drosophila) |
| chr1_+_117452669 | 0.22 |
ENST00000393203.2
|
PTGFRN
|
prostaglandin F2 receptor inhibitor |
| chr1_-_120612240 | 0.21 |
ENST00000256646.2
|
NOTCH2
|
notch 2 |
| chr1_+_40862501 | 0.21 |
ENST00000539317.1
|
SMAP2
|
small ArfGAP2 |
| chr2_+_46926048 | 0.21 |
ENST00000306503.5
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr2_+_235860616 | 0.21 |
ENST00000392011.2
|
SH3BP4
|
SH3-domain binding protein 4 |
| chr1_-_38325256 | 0.20 |
ENST00000373036.4
|
MTF1
|
metal-regulatory transcription factor 1 |
| chr2_+_173940442 | 0.20 |
ENST00000409176.2
ENST00000338983.3 ENST00000431503.2 |
MLTK
|
Mitogen-activated protein kinase kinase kinase MLT |
| chr2_+_36582857 | 0.20 |
ENST00000280527.2
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
| chr9_+_125703282 | 0.19 |
ENST00000373647.4
ENST00000402311.1 |
RABGAP1
|
RAB GTPase activating protein 1 |
| chr18_-_45456930 | 0.19 |
ENST00000262160.6
ENST00000587269.1 |
SMAD2
|
SMAD family member 2 |
| chr7_-_121036337 | 0.19 |
ENST00000426156.1
ENST00000359943.3 ENST00000412653.1 |
FAM3C
|
family with sequence similarity 3, member C |
| chr16_-_85722530 | 0.18 |
ENST00000253462.3
|
GINS2
|
GINS complex subunit 2 (Psf2 homolog) |
| chr3_+_107241783 | 0.18 |
ENST00000415149.2
ENST00000402543.1 ENST00000325805.8 ENST00000427402.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr1_+_32573636 | 0.18 |
ENST00000373625.3
|
KPNA6
|
karyopherin alpha 6 (importin alpha 7) |
| chr17_-_40306934 | 0.17 |
ENST00000592574.1
ENST00000550406.1 ENST00000547517.1 ENST00000393860.3 ENST00000346213.4 |
CTD-2132N18.3
RAB5C
|
Uncharacterized protein RAB5C, member RAS oncogene family |
| chr2_-_153032484 | 0.17 |
ENST00000263904.4
|
STAM2
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 |
| chr18_+_20513782 | 0.16 |
ENST00000399722.2
ENST00000399725.2 ENST00000399721.2 ENST00000583594.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr3_+_42695176 | 0.15 |
ENST00000232974.6
ENST00000457842.3 |
ZBTB47
|
zinc finger and BTB domain containing 47 |
| chr1_+_162467595 | 0.15 |
ENST00000538489.1
ENST00000489294.1 |
UHMK1
|
U2AF homology motif (UHM) kinase 1 |
| chr20_+_43595115 | 0.14 |
ENST00000372806.3
ENST00000396731.4 ENST00000372801.1 ENST00000499879.2 |
STK4
|
serine/threonine kinase 4 |
| chr5_-_171711061 | 0.14 |
ENST00000393792.2
|
UBTD2
|
ubiquitin domain containing 2 |
| chr3_+_37903432 | 0.14 |
ENST00000443503.2
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr18_-_29522989 | 0.14 |
ENST00000582539.1
ENST00000283351.4 ENST00000582513.1 |
TRAPPC8
|
trafficking protein particle complex 8 |
| chr11_-_116968987 | 0.13 |
ENST00000434315.2
ENST00000292055.4 ENST00000375288.1 ENST00000542607.1 ENST00000445177.1 ENST00000375300.1 ENST00000446921.2 |
SIK3
|
SIK family kinase 3 |
| chr17_-_62340581 | 0.12 |
ENST00000258991.3
ENST00000583738.1 ENST00000584379.1 |
TEX2
|
testis expressed 2 |
| chr12_-_44200052 | 0.12 |
ENST00000548315.1
ENST00000552521.1 ENST00000546662.1 ENST00000548403.1 ENST00000546506.1 |
TWF1
|
twinfilin actin-binding protein 1 |
| chr12_+_19592602 | 0.12 |
ENST00000398864.3
ENST00000266508.9 |
AEBP2
|
AE binding protein 2 |
| chr21_-_36260980 | 0.12 |
ENST00000344691.4
ENST00000358356.5 |
RUNX1
|
runt-related transcription factor 1 |
| chr9_+_91003271 | 0.12 |
ENST00000375859.3
ENST00000541629.1 |
SPIN1
|
spindlin 1 |
| chr5_-_142065612 | 0.12 |
ENST00000360966.5
ENST00000411960.1 |
FGF1
|
fibroblast growth factor 1 (acidic) |
| chr7_-_111846435 | 0.12 |
ENST00000437633.1
ENST00000428084.1 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr8_-_18871159 | 0.12 |
ENST00000327040.8
ENST00000440756.2 |
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr1_+_201617450 | 0.11 |
ENST00000295624.6
ENST00000367297.4 ENST00000367300.3 |
NAV1
|
neuron navigator 1 |
| chr4_+_183164574 | 0.11 |
ENST00000511685.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr5_+_78532003 | 0.11 |
ENST00000396137.4
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chr3_+_105085734 | 0.11 |
ENST00000306107.5
|
ALCAM
|
activated leukocyte cell adhesion molecule |
| chr11_+_94822968 | 0.10 |
ENST00000278505.4
|
ENDOD1
|
endonuclease domain containing 1 |
| chr8_+_8860314 | 0.10 |
ENST00000250263.7
ENST00000519292.1 |
ERI1
|
exoribonuclease 1 |
| chr17_+_27717415 | 0.10 |
ENST00000583121.1
ENST00000261716.3 |
TAOK1
|
TAO kinase 1 |
| chr5_-_43313574 | 0.09 |
ENST00000325110.6
ENST00000433297.2 |
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr22_+_38093005 | 0.09 |
ENST00000406386.3
|
TRIOBP
|
TRIO and F-actin binding protein |
| chr10_-_119806085 | 0.09 |
ENST00000355624.3
|
RAB11FIP2
|
RAB11 family interacting protein 2 (class I) |
| chr5_-_169407744 | 0.09 |
ENST00000377365.3
|
FAM196B
|
family with sequence similarity 196, member B |
| chr6_-_111136513 | 0.09 |
ENST00000368911.3
|
CDK19
|
cyclin-dependent kinase 19 |
| chr17_+_46985731 | 0.09 |
ENST00000360943.5
|
UBE2Z
|
ubiquitin-conjugating enzyme E2Z |
| chr5_-_73937244 | 0.09 |
ENST00000302351.4
ENST00000510316.1 ENST00000508331.1 |
ENC1
|
ectodermal-neural cortex 1 (with BTB domain) |
| chr20_+_56884752 | 0.09 |
ENST00000244040.3
|
RAB22A
|
RAB22A, member RAS oncogene family |
| chr12_+_109535373 | 0.09 |
ENST00000242576.2
|
UNG
|
uracil-DNA glycosylase |
| chr5_+_125935960 | 0.08 |
ENST00000297540.4
|
PHAX
|
phosphorylated adaptor for RNA export |
| chr9_-_123639600 | 0.08 |
ENST00000373896.3
|
PHF19
|
PHD finger protein 19 |
| chr1_+_78245303 | 0.08 |
ENST00000370791.3
ENST00000443751.2 |
FAM73A
|
family with sequence similarity 73, member A |
| chr5_+_108083517 | 0.08 |
ENST00000281092.4
ENST00000536402.1 |
FER
|
fer (fps/fes related) tyrosine kinase |
| chr4_+_79697495 | 0.08 |
ENST00000502871.1
ENST00000335016.5 |
BMP2K
|
BMP2 inducible kinase |
| chr3_+_152017181 | 0.07 |
ENST00000498502.1
ENST00000324196.5 ENST00000545754.1 ENST00000357472.3 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr1_+_62208091 | 0.07 |
ENST00000316485.6
ENST00000371158.2 |
INADL
|
InaD-like (Drosophila) |
| chr11_-_34379546 | 0.07 |
ENST00000435224.2
|
ABTB2
|
ankyrin repeat and BTB (POZ) domain containing 2 |
| chr3_-_169899504 | 0.07 |
ENST00000474275.1
ENST00000484931.1 ENST00000494943.1 ENST00000497658.1 ENST00000465896.1 ENST00000475729.1 ENST00000495893.2 ENST00000481639.1 ENST00000467570.1 ENST00000466189.1 |
PHC3
|
polyhomeotic homolog 3 (Drosophila) |
| chr1_-_228135599 | 0.07 |
ENST00000272164.5
|
WNT9A
|
wingless-type MMTV integration site family, member 9A |
| chr3_-_185542817 | 0.07 |
ENST00000382199.2
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr15_-_56209306 | 0.07 |
ENST00000506154.1
ENST00000338963.2 ENST00000508342.1 |
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
| chr1_+_22379120 | 0.06 |
ENST00000400259.1
ENST00000344548.3 |
CDC42
|
cell division cycle 42 |
| chr9_+_116638562 | 0.06 |
ENST00000374126.5
ENST00000288466.7 |
ZNF618
|
zinc finger protein 618 |
| chr17_-_1395954 | 0.06 |
ENST00000359786.5
|
MYO1C
|
myosin IC |
| chr8_+_28747884 | 0.06 |
ENST00000287701.10
ENST00000444075.1 ENST00000403668.2 ENST00000519662.1 ENST00000558662.1 ENST00000523613.1 ENST00000560599.1 ENST00000397358.3 |
HMBOX1
|
homeobox containing 1 |
| chr3_+_171758344 | 0.06 |
ENST00000336824.4
ENST00000423424.1 |
FNDC3B
|
fibronectin type III domain containing 3B |
| chr10_-_735553 | 0.06 |
ENST00000280886.6
ENST00000423550.1 |
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
| chr1_-_95007193 | 0.06 |
ENST00000370207.4
ENST00000334047.7 |
F3
|
coagulation factor III (thromboplastin, tissue factor) |
| chr10_-_71930222 | 0.06 |
ENST00000458634.2
ENST00000373239.2 ENST00000373242.2 ENST00000373241.4 |
SAR1A
|
SAR1 homolog A (S. cerevisiae) |
| chr6_-_86352642 | 0.06 |
ENST00000355238.6
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr2_+_24714729 | 0.05 |
ENST00000406961.1
ENST00000405141.1 |
NCOA1
|
nuclear receptor coactivator 1 |
| chr10_+_72575643 | 0.05 |
ENST00000373202.3
|
SGPL1
|
sphingosine-1-phosphate lyase 1 |
| chr4_+_6784401 | 0.05 |
ENST00000425103.1
ENST00000307659.5 |
KIAA0232
|
KIAA0232 |
| chr11_+_109964087 | 0.05 |
ENST00000278590.3
|
ZC3H12C
|
zinc finger CCCH-type containing 12C |
| chr3_-_33481835 | 0.05 |
ENST00000283629.3
|
UBP1
|
upstream binding protein 1 (LBP-1a) |
| chr17_-_7382834 | 0.05 |
ENST00000380599.4
|
ZBTB4
|
zinc finger and BTB domain containing 4 |
| chrX_-_24045303 | 0.04 |
ENST00000328046.8
|
KLHL15
|
kelch-like family member 15 |
| chr5_-_78809950 | 0.04 |
ENST00000334082.6
|
HOMER1
|
homer homolog 1 (Drosophila) |
| chr11_-_79151695 | 0.04 |
ENST00000278550.7
|
TENM4
|
teneurin transmembrane protein 4 |
| chr6_-_53409890 | 0.04 |
ENST00000229416.6
|
GCLC
|
glutamate-cysteine ligase, catalytic subunit |
| chr5_+_96271141 | 0.04 |
ENST00000231368.5
|
LNPEP
|
leucyl/cystinyl aminopeptidase |
| chr9_+_140513438 | 0.04 |
ENST00000462484.1
ENST00000334856.6 ENST00000460843.1 |
EHMT1
|
euchromatic histone-lysine N-methyltransferase 1 |
| chr3_+_141205852 | 0.03 |
ENST00000286364.3
ENST00000452898.1 |
RASA2
|
RAS p21 protein activator 2 |
| chr4_+_154125565 | 0.03 |
ENST00000338700.5
|
TRIM2
|
tripartite motif containing 2 |
| chr1_+_100435315 | 0.03 |
ENST00000370155.3
ENST00000465289.1 |
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
| chr4_-_157892498 | 0.03 |
ENST00000502773.1
|
PDGFC
|
platelet derived growth factor C |
| chr20_+_43104508 | 0.03 |
ENST00000262605.4
ENST00000372904.3 |
TTPAL
|
tocopherol (alpha) transfer protein-like |
| chr6_+_122720681 | 0.03 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chrX_+_108780062 | 0.03 |
ENST00000372106.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr20_+_30865429 | 0.03 |
ENST00000375712.3
|
KIF3B
|
kinesin family member 3B |
| chr22_-_44258360 | 0.03 |
ENST00000330884.4
ENST00000249130.5 |
SULT4A1
|
sulfotransferase family 4A, member 1 |
| chr1_-_55680762 | 0.03 |
ENST00000407756.1
ENST00000294383.6 |
USP24
|
ubiquitin specific peptidase 24 |
| chr6_+_134274322 | 0.03 |
ENST00000367871.1
ENST00000237264.4 |
TBPL1
|
TBP-like 1 |
| chrX_-_80065146 | 0.03 |
ENST00000373275.4
|
BRWD3
|
bromodomain and WD repeat domain containing 3 |
| chr10_-_98273668 | 0.03 |
ENST00000357947.3
|
TLL2
|
tolloid-like 2 |
| chr2_-_201828356 | 0.02 |
ENST00000234296.2
|
ORC2
|
origin recognition complex, subunit 2 |
| chr4_-_185395672 | 0.02 |
ENST00000393593.3
|
IRF2
|
interferon regulatory factor 2 |
| chr6_-_11232891 | 0.02 |
ENST00000379433.5
ENST00000379446.5 |
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr3_+_43328004 | 0.02 |
ENST00000454177.1
ENST00000429705.2 ENST00000296088.7 ENST00000437827.1 |
SNRK
|
SNF related kinase |
| chr5_-_142783175 | 0.02 |
ENST00000231509.3
ENST00000394464.2 |
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chrX_-_19905703 | 0.02 |
ENST00000397821.3
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr9_+_15553055 | 0.02 |
ENST00000380701.3
|
CCDC171
|
coiled-coil domain containing 171 |
| chr10_+_102295616 | 0.01 |
ENST00000299163.6
|
HIF1AN
|
hypoxia inducible factor 1, alpha subunit inhibitor |
| chr16_-_74640986 | 0.01 |
ENST00000422840.2
ENST00000565260.1 ENST00000447066.2 ENST00000205061.5 |
GLG1
|
golgi glycoprotein 1 |
| chr4_+_25314388 | 0.01 |
ENST00000302874.4
|
ZCCHC4
|
zinc finger, CCHC domain containing 4 |
| chr5_-_180288248 | 0.01 |
ENST00000512132.1
ENST00000506439.1 ENST00000502412.1 ENST00000359141.6 |
ZFP62
|
ZFP62 zinc finger protein |
| chr1_-_109506036 | 0.01 |
ENST00000369976.1
ENST00000356970.2 ENST00000369971.2 ENST00000415331.1 ENST00000357393.4 |
CLCC1
AKNAD1
|
chloride channel CLIC-like 1 AKNA domain containing 1 |
| chr21_-_40685477 | 0.01 |
ENST00000342449.3
|
BRWD1
|
bromodomain and WD repeat domain containing 1 |
| chr7_+_129074266 | 0.01 |
ENST00000249344.2
ENST00000435494.2 |
STRIP2
|
striatin interacting protein 2 |
| chr12_-_56652111 | 0.01 |
ENST00000267116.7
|
ANKRD52
|
ankyrin repeat domain 52 |
| chr12_-_56583332 | 0.01 |
ENST00000347471.4
ENST00000267064.4 ENST00000394023.3 |
SMARCC2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
| chr7_+_90225796 | 0.01 |
ENST00000380050.3
|
CDK14
|
cyclin-dependent kinase 14 |
| chr12_+_56360550 | 0.01 |
ENST00000266970.4
|
CDK2
|
cyclin-dependent kinase 2 |
| chr8_-_81787006 | 0.01 |
ENST00000327835.3
|
ZNF704
|
zinc finger protein 704 |
| chr14_+_55518349 | 0.01 |
ENST00000395468.4
|
MAPK1IP1L
|
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr19_+_13229126 | 0.01 |
ENST00000292431.4
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr12_+_50690489 | 0.01 |
ENST00000598429.1
|
AC140061.12
|
Uncharacterized protein |
| chr1_+_84543734 | 0.00 |
ENST00000370689.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr1_-_110052302 | 0.00 |
ENST00000369864.4
ENST00000369862.1 |
AMIGO1
|
adhesion molecule with Ig-like domain 1 |
| chr2_+_28615669 | 0.00 |
ENST00000379619.1
ENST00000264716.4 |
FOSL2
|
FOS-like antigen 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of AAGGUGC
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0002605 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) negative regulation of dendritic cell antigen processing and presentation(GO:0002605) negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.1 | 0.3 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.1 | 0.3 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.3 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.2 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.2 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.2 | GO:0030047 | actin modification(GO:0030047) |
| 0.0 | 0.3 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.1 | GO:0050904 | diapedesis(GO:0050904) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.3 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.1 | GO:0006408 | snRNA export from nucleus(GO:0006408) |
| 0.0 | 0.2 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.1 | GO:0045925 | positive regulation of female receptivity(GO:0045925) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.0 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.3 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.1 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.0 | 0.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.1 | GO:0090135 | positive regulation of synapse structural plasticity(GO:0051835) actin filament branching(GO:0090135) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.0 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.2 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.2 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.3 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.3 | GO:0019961 | interferon binding(GO:0019961) |
| 0.1 | 0.3 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.1 | 0.2 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.0 | 0.1 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.0 | 0.3 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.3 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.3 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.2 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.3 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |