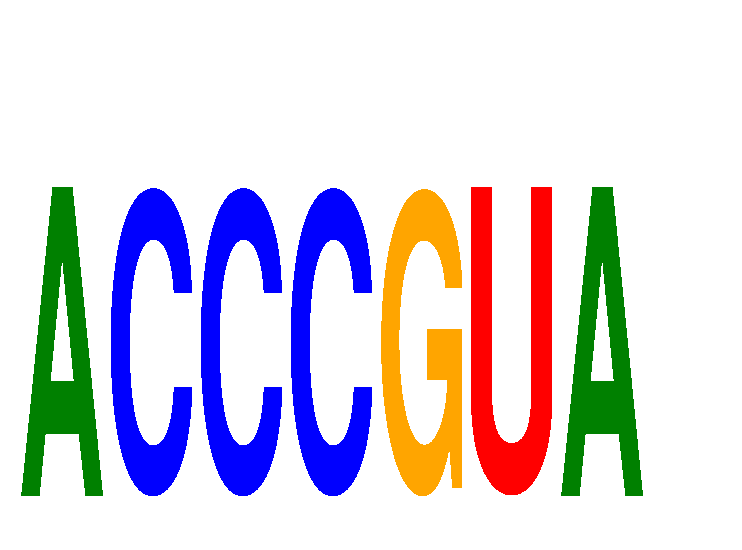Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for ACCCGUA
Z-value: 0.03
miRNA associated with seed ACCCGUA
| Name | miRBASE accession |
|---|---|
|
hsa-miR-99a-5p
|
MIMAT0000097 |
|
hsa-miR-99b-5p
|
MIMAT0000689 |
|
hsa-miR-100-5p
|
MIMAT0000098 |
Activity profile of ACCCGUA motif
Sorted Z-values of ACCCGUA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_35930219 | 0.02 |
ENST00000374694.1
|
FZD8
|
frizzled family receptor 8 |
| chr2_-_72375167 | 0.02 |
ENST00000001146.2
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1 |
| chr8_+_126442563 | 0.01 |
ENST00000311922.3
|
TRIB1
|
tribbles pseudokinase 1 |
| chr8_-_141645645 | 0.01 |
ENST00000519980.1
ENST00000220592.5 |
AGO2
|
argonaute RISC catalytic component 2 |
| chr3_+_32859510 | 0.01 |
ENST00000383763.5
|
TRIM71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr2_-_208634287 | 0.01 |
ENST00000295417.3
|
FZD5
|
frizzled family receptor 5 |
| chr2_+_33661382 | 0.01 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr6_-_90121938 | 0.00 |
ENST00000369415.4
|
RRAGD
|
Ras-related GTP binding D |
| chr17_-_4046257 | 0.00 |
ENST00000381638.2
|
ZZEF1
|
zinc finger, ZZ-type with EF-hand domain 1 |
| chr8_+_75736761 | 0.00 |
ENST00000260113.2
|
PI15
|
peptidase inhibitor 15 |
| chr20_+_20348740 | 0.00 |
ENST00000310227.1
|
INSM1
|
insulinoma-associated 1 |
| chr7_+_30323923 | 0.00 |
ENST00000323037.4
|
ZNRF2
|
zinc and ring finger 2 |
| chr3_+_152017181 | 0.00 |
ENST00000498502.1
ENST00000324196.5 ENST00000545754.1 ENST00000357472.3 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr2_+_86947296 | 0.00 |
ENST00000283632.4
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae) |
| chr1_+_244816237 | 0.00 |
ENST00000302550.11
|
DESI2
|
desumoylating isopeptidase 2 |
| chr11_+_86748863 | 0.00 |
ENST00000340353.7
|
TMEM135
|
transmembrane protein 135 |