Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
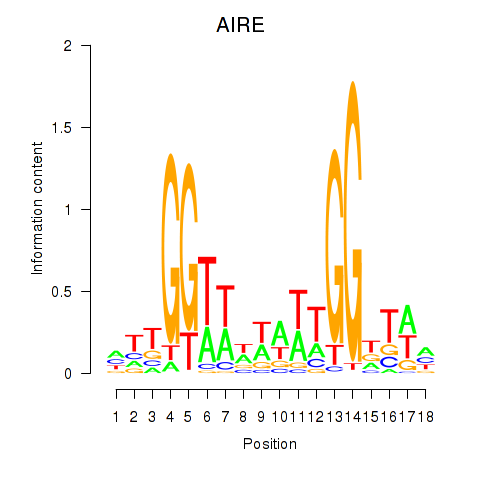
Results for AIRE
Z-value: 0.33
Transcription factors associated with AIRE
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
AIRE
|
ENSG00000160224.12 | autoimmune regulator |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| AIRE | hg19_v2_chr21_+_45705752_45705767 | -0.09 | 7.0e-01 | Click! |
Activity profile of AIRE motif
Sorted Z-values of AIRE motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_31147020 | 0.59 |
ENST00000568261.1
ENST00000567797.1 ENST00000317508.6 |
PRSS8
|
protease, serine, 8 |
| chr1_+_79115503 | 0.58 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chrM_+_12331 | 0.57 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chrM_-_14670 | 0.57 |
ENST00000361681.2
|
MT-ND6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr11_-_327537 | 0.55 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr19_-_52551814 | 0.48 |
ENST00000594154.1
ENST00000598745.1 ENST00000597273.1 |
ZNF432
|
zinc finger protein 432 |
| chr3_+_189507460 | 0.46 |
ENST00000434928.1
|
TP63
|
tumor protein p63 |
| chr12_+_75874580 | 0.45 |
ENST00000456650.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr7_-_87936195 | 0.45 |
ENST00000414498.1
ENST00000301959.5 ENST00000380079.4 |
STEAP4
|
STEAP family member 4 |
| chr11_+_35211511 | 0.44 |
ENST00000524922.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr19_-_52552051 | 0.44 |
ENST00000221315.5
|
ZNF432
|
zinc finger protein 432 |
| chr22_+_36649170 | 0.43 |
ENST00000438034.1
ENST00000427990.1 ENST00000347595.7 ENST00000397279.4 ENST00000433768.1 ENST00000440669.2 |
APOL1
|
apolipoprotein L, 1 |
| chr12_+_75874984 | 0.42 |
ENST00000550491.1
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr4_-_120222076 | 0.42 |
ENST00000504110.1
|
C4orf3
|
chromosome 4 open reading frame 3 |
| chrM_+_14741 | 0.35 |
ENST00000361789.2
|
MT-CYB
|
mitochondrially encoded cytochrome b |
| chr22_+_36649056 | 0.35 |
ENST00000397278.3
ENST00000422706.1 ENST00000426053.1 ENST00000319136.4 |
APOL1
|
apolipoprotein L, 1 |
| chr12_-_50677255 | 0.34 |
ENST00000551691.1
ENST00000394943.3 ENST00000341247.4 |
LIMA1
|
LIM domain and actin binding 1 |
| chrM_+_4431 | 0.34 |
ENST00000361453.3
|
MT-ND2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chrM_+_7586 | 0.34 |
ENST00000361739.1
|
MT-CO2
|
mitochondrially encoded cytochrome c oxidase II |
| chr16_-_31146961 | 0.33 |
ENST00000567531.1
|
PRSS8
|
protease, serine, 8 |
| chr20_+_44637526 | 0.33 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chrM_+_8366 | 0.30 |
ENST00000361851.1
|
MT-ATP8
|
mitochondrially encoded ATP synthase 8 |
| chr21_+_39644305 | 0.30 |
ENST00000398930.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr21_+_39644172 | 0.29 |
ENST00000398932.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr21_+_39644395 | 0.29 |
ENST00000398934.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr21_+_39644214 | 0.25 |
ENST00000438657.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr9_-_21482312 | 0.25 |
ENST00000448696.3
|
IFNE
|
interferon, epsilon |
| chr20_+_12989596 | 0.25 |
ENST00000434210.1
ENST00000399002.2 |
SPTLC3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr10_-_4720333 | 0.24 |
ENST00000430998.2
|
LINC00704
|
long intergenic non-protein coding RNA 704 |
| chr12_+_7282795 | 0.22 |
ENST00000266546.6
|
CLSTN3
|
calsyntenin 3 |
| chr19_-_21950362 | 0.21 |
ENST00000358296.6
|
ZNF100
|
zinc finger protein 100 |
| chr19_-_36304201 | 0.21 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr13_-_46756351 | 0.21 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr19_+_11455900 | 0.20 |
ENST00000588790.1
|
CCDC159
|
coiled-coil domain containing 159 |
| chr12_+_75874460 | 0.20 |
ENST00000266659.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr15_+_52043758 | 0.19 |
ENST00000249700.4
ENST00000539962.2 |
TMOD2
|
tropomodulin 2 (neuronal) |
| chr1_+_174843548 | 0.18 |
ENST00000478442.1
ENST00000465412.1 |
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr19_-_23869999 | 0.18 |
ENST00000601935.1
ENST00000359788.4 ENST00000600313.1 ENST00000596211.1 ENST00000599168.1 |
ZNF675
|
zinc finger protein 675 |
| chr6_-_33266687 | 0.18 |
ENST00000444031.2
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr10_+_127661942 | 0.18 |
ENST00000417114.1
ENST00000445510.1 ENST00000368691.1 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr7_-_64467031 | 0.18 |
ENST00000394323.2
|
ERV3-1
|
endogenous retrovirus group 3, member 1 |
| chr4_-_186696425 | 0.17 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr16_-_29499154 | 0.17 |
ENST00000354563.5
|
RP11-231C14.4
|
Uncharacterized protein |
| chr7_+_116451100 | 0.16 |
ENST00000464223.1
ENST00000484325.1 |
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr4_+_69917078 | 0.15 |
ENST00000502942.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chrM_+_5824 | 0.15 |
ENST00000361624.2
|
MT-CO1
|
mitochondrially encoded cytochrome c oxidase I |
| chr19_-_23869970 | 0.15 |
ENST00000601010.1
|
ZNF675
|
zinc finger protein 675 |
| chr12_-_59175485 | 0.14 |
ENST00000550678.1
ENST00000552201.1 |
RP11-767I20.1
|
RP11-767I20.1 |
| chr4_-_57524061 | 0.14 |
ENST00000508121.1
|
HOPX
|
HOP homeobox |
| chr19_+_16607122 | 0.14 |
ENST00000221671.3
ENST00000594035.1 ENST00000599550.1 ENST00000594813.1 |
C19orf44
|
chromosome 19 open reading frame 44 |
| chr5_+_667759 | 0.14 |
ENST00000594226.1
|
AC026740.1
|
Uncharacterized protein |
| chr4_+_71587669 | 0.14 |
ENST00000381006.3
ENST00000226328.4 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr1_+_40840320 | 0.13 |
ENST00000372708.1
|
SMAP2
|
small ArfGAP2 |
| chr4_+_71588372 | 0.13 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr19_-_36643329 | 0.13 |
ENST00000589154.1
|
COX7A1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr13_-_26795840 | 0.13 |
ENST00000381570.3
ENST00000399762.2 ENST00000346166.3 |
RNF6
|
ring finger protein (C3H2C3 type) 6 |
| chr1_-_35497506 | 0.13 |
ENST00000317538.5
ENST00000373340.2 ENST00000357182.4 |
ZMYM6
|
zinc finger, MYM-type 6 |
| chr22_-_32767017 | 0.12 |
ENST00000400234.1
|
RFPL3S
|
RFPL3 antisense |
| chr15_+_52043813 | 0.12 |
ENST00000435126.2
|
TMOD2
|
tropomodulin 2 (neuronal) |
| chr12_-_113772835 | 0.12 |
ENST00000552014.1
ENST00000548186.1 ENST00000202831.3 ENST00000549181.1 |
SLC8B1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr10_-_75682535 | 0.12 |
ENST00000409178.1
|
C10orf55
|
chromosome 10 open reading frame 55 |
| chr1_+_36038971 | 0.12 |
ENST00000373235.3
|
TFAP2E
|
transcription factor AP-2 epsilon (activating enhancer binding protein 2 epsilon) |
| chr9_+_71986182 | 0.12 |
ENST00000303068.7
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr21_+_30502806 | 0.12 |
ENST00000399928.1
ENST00000399926.1 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr16_+_14802801 | 0.11 |
ENST00000526520.1
ENST00000531598.2 |
NPIPA3
|
nuclear pore complex interacting protein family, member A3 |
| chr15_-_83209210 | 0.11 |
ENST00000561157.1
ENST00000330244.6 |
RPS17L
|
ribosomal protein S17-like |
| chr8_+_128748308 | 0.11 |
ENST00000377970.2
|
MYC
|
v-myc avian myelocytomatosis viral oncogene homolog |
| chr6_+_22221010 | 0.10 |
ENST00000567753.1
|
RP11-524C21.2
|
RP11-524C21.2 |
| chr1_-_52344471 | 0.10 |
ENST00000352171.7
ENST00000354831.7 |
NRD1
|
nardilysin (N-arginine dibasic convertase) |
| chr12_+_55248289 | 0.10 |
ENST00000308796.6
|
MUCL1
|
mucin-like 1 |
| chr7_+_142374104 | 0.10 |
ENST00000604952.1
|
MTRNR2L6
|
MT-RNR2-like 6 |
| chr19_-_16606988 | 0.09 |
ENST00000269881.3
|
CALR3
|
calreticulin 3 |
| chr4_-_16085657 | 0.09 |
ENST00000543373.1
|
PROM1
|
prominin 1 |
| chr9_-_25678856 | 0.09 |
ENST00000358022.3
|
TUSC1
|
tumor suppressor candidate 1 |
| chr6_+_42584847 | 0.09 |
ENST00000372883.3
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr4_+_1385340 | 0.09 |
ENST00000324803.4
|
CRIPAK
|
cysteine-rich PAK1 inhibitor |
| chr19_-_16606930 | 0.09 |
ENST00000600762.1
|
CALR3
|
calreticulin 3 |
| chr10_+_115511213 | 0.09 |
ENST00000361048.1
|
PLEKHS1
|
pleckstrin homology domain containing, family S member 1 |
| chr3_-_121468602 | 0.09 |
ENST00000340645.5
|
GOLGB1
|
golgin B1 |
| chr20_+_58203664 | 0.08 |
ENST00000541461.1
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr2_+_219840955 | 0.08 |
ENST00000598002.1
ENST00000432733.1 |
LINC00608
|
long intergenic non-protein coding RNA 608 |
| chr12_-_64784471 | 0.08 |
ENST00000333722.5
|
C12orf56
|
chromosome 12 open reading frame 56 |
| chr5_-_38845812 | 0.08 |
ENST00000513480.1
ENST00000512519.1 |
CTD-2127H9.1
|
CTD-2127H9.1 |
| chr18_+_39739223 | 0.08 |
ENST00000601948.1
|
LINC00907
|
long intergenic non-protein coding RNA 907 |
| chr20_+_44036900 | 0.08 |
ENST00000443296.1
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr6_-_32806506 | 0.07 |
ENST00000374897.2
ENST00000452392.2 |
TAP2
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) Uncharacterized protein |
| chr20_+_44036620 | 0.07 |
ENST00000372710.3
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr19_+_18208603 | 0.07 |
ENST00000262811.6
|
MAST3
|
microtubule associated serine/threonine kinase 3 |
| chr11_+_107650219 | 0.07 |
ENST00000398067.1
|
AP001024.1
|
Uncharacterized protein |
| chr11_-_26744908 | 0.07 |
ENST00000533617.1
|
SLC5A12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr9_+_117904097 | 0.07 |
ENST00000374016.1
|
DEC1
|
deleted in esophageal cancer 1 |
| chr17_+_41857793 | 0.07 |
ENST00000449302.3
|
C17orf105
|
chromosome 17 open reading frame 105 |
| chr3_-_176914191 | 0.07 |
ENST00000437738.1
ENST00000424913.1 ENST00000443315.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr1_+_174670143 | 0.07 |
ENST00000367687.1
ENST00000347255.2 |
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr14_-_24911971 | 0.07 |
ENST00000555365.1
ENST00000399395.3 |
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr2_+_179316163 | 0.06 |
ENST00000409117.3
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr1_+_207669573 | 0.06 |
ENST00000400960.2
ENST00000534202.1 |
CR1
|
complement component (3b/4b) receptor 1 (Knops blood group) |
| chr4_-_4544061 | 0.06 |
ENST00000507908.1
|
STX18
|
syntaxin 18 |
| chr19_-_4902877 | 0.06 |
ENST00000381781.2
|
ARRDC5
|
arrestin domain containing 5 |
| chr3_-_121468513 | 0.06 |
ENST00000494517.1
ENST00000393667.3 |
GOLGB1
|
golgin B1 |
| chr1_+_207669613 | 0.06 |
ENST00000367049.4
ENST00000529814.1 |
CR1
|
complement component (3b/4b) receptor 1 (Knops blood group) |
| chr19_-_47249679 | 0.06 |
ENST00000263280.6
|
STRN4
|
striatin, calmodulin binding protein 4 |
| chr1_-_152297679 | 0.06 |
ENST00000368799.1
|
FLG
|
filaggrin |
| chr12_+_12202785 | 0.06 |
ENST00000586576.1
ENST00000464885.2 |
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chr10_-_13350447 | 0.06 |
ENST00000429930.1
|
AL138764.1
|
Uncharacterized protein |
| chr2_+_204732487 | 0.05 |
ENST00000302823.3
|
CTLA4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr10_+_115511434 | 0.05 |
ENST00000369312.4
|
PLEKHS1
|
pleckstrin homology domain containing, family S member 1 |
| chr14_-_24912047 | 0.05 |
ENST00000553930.1
|
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr19_+_53517174 | 0.05 |
ENST00000602168.1
|
ERVV-1
|
endogenous retrovirus group V, member 1 |
| chr1_+_96208646 | 0.05 |
ENST00000603401.1
|
RP11-286B14.2
|
RP11-286B14.2 |
| chr7_+_80255472 | 0.05 |
ENST00000428497.1
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr12_+_48516463 | 0.05 |
ENST00000546465.1
|
PFKM
|
phosphofructokinase, muscle |
| chr5_+_122181184 | 0.05 |
ENST00000513881.1
|
SNX24
|
sorting nexin 24 |
| chr4_-_88450535 | 0.05 |
ENST00000541496.1
|
SPARCL1
|
SPARC-like 1 (hevin) |
| chr19_-_10491234 | 0.05 |
ENST00000524462.1
ENST00000531836.1 ENST00000525621.1 |
TYK2
|
tyrosine kinase 2 |
| chr2_+_38177620 | 0.05 |
ENST00000402091.3
|
RMDN2
|
regulator of microtubule dynamics 2 |
| chr17_-_39150385 | 0.05 |
ENST00000391586.1
|
KRTAP3-3
|
keratin associated protein 3-3 |
| chr9_-_70429731 | 0.05 |
ENST00000377413.1
|
FOXD4L4
|
forkhead box D4-like 4 |
| chr16_+_50914085 | 0.04 |
ENST00000576842.1
|
CTD-2034I21.2
|
CTD-2034I21.2 |
| chr11_+_45376922 | 0.04 |
ENST00000524410.1
ENST00000524488.1 ENST00000524565.1 |
RP11-430H10.1
|
RP11-430H10.1 |
| chrX_-_49041242 | 0.04 |
ENST00000453382.1
ENST00000540849.1 ENST00000536904.1 ENST00000432913.1 |
PRICKLE3
|
prickle homolog 3 (Drosophila) |
| chrX_-_153191674 | 0.04 |
ENST00000350060.5
ENST00000370016.1 |
ARHGAP4
|
Rho GTPase activating protein 4 |
| chr19_-_59084922 | 0.04 |
ENST00000215057.2
ENST00000599369.1 |
MZF1
|
myeloid zinc finger 1 |
| chr14_-_46185155 | 0.04 |
ENST00000555442.1
|
RP11-369C8.1
|
RP11-369C8.1 |
| chr10_+_82300575 | 0.04 |
ENST00000313455.4
|
SH2D4B
|
SH2 domain containing 4B |
| chr11_+_6947647 | 0.04 |
ENST00000278319.5
|
ZNF215
|
zinc finger protein 215 |
| chr19_+_56751773 | 0.04 |
ENST00000600684.1
|
ZSCAN5D
|
zinc finger and SCAN domain containing 5D |
| chr8_+_104384616 | 0.04 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr2_+_207804278 | 0.04 |
ENST00000272852.3
|
CPO
|
carboxypeptidase O |
| chr6_+_167525277 | 0.04 |
ENST00000400926.2
|
CCR6
|
chemokine (C-C motif) receptor 6 |
| chr6_+_168434678 | 0.04 |
ENST00000496008.1
|
KIF25
|
kinesin family member 25 |
| chrX_-_153191708 | 0.04 |
ENST00000393721.1
ENST00000370028.3 |
ARHGAP4
|
Rho GTPase activating protein 4 |
| chr2_-_208994548 | 0.04 |
ENST00000282141.3
|
CRYGC
|
crystallin, gamma C |
| chr16_-_56223480 | 0.04 |
ENST00000565155.1
|
RP11-461O7.1
|
RP11-461O7.1 |
| chr17_+_34171081 | 0.04 |
ENST00000585577.1
|
TAF15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa |
| chr2_-_127977654 | 0.04 |
ENST00000409327.1
|
CYP27C1
|
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr3_-_123710199 | 0.04 |
ENST00000184183.4
|
ROPN1
|
rhophilin associated tail protein 1 |
| chr1_-_167883327 | 0.04 |
ENST00000476818.2
ENST00000367851.4 ENST00000367848.1 |
ADCY10
|
adenylate cyclase 10 (soluble) |
| chr7_-_36764004 | 0.04 |
ENST00000431169.1
|
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr1_+_241815577 | 0.04 |
ENST00000366552.2
ENST00000437684.2 |
WDR64
|
WD repeat domain 64 |
| chr10_-_101841588 | 0.03 |
ENST00000370418.3
|
CPN1
|
carboxypeptidase N, polypeptide 1 |
| chr4_+_76649797 | 0.03 |
ENST00000538159.1
ENST00000514213.2 |
USO1
|
USO1 vesicle transport factor |
| chr2_-_166060571 | 0.03 |
ENST00000360093.3
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr17_+_3323862 | 0.03 |
ENST00000291231.1
|
OR3A3
|
olfactory receptor, family 3, subfamily A, member 3 |
| chr2_+_166150541 | 0.03 |
ENST00000283256.6
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr8_+_24241789 | 0.03 |
ENST00000256412.4
ENST00000538205.1 |
ADAMDEC1
|
ADAM-like, decysin 1 |
| chr20_-_43150601 | 0.03 |
ENST00000541235.1
ENST00000255175.1 ENST00000342374.4 |
SERINC3
|
serine incorporator 3 |
| chr5_+_122181279 | 0.03 |
ENST00000395451.4
ENST00000506996.1 |
SNX24
|
sorting nexin 24 |
| chr12_+_12202774 | 0.03 |
ENST00000589718.1
|
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chr15_-_81202118 | 0.03 |
ENST00000560560.1
|
RP11-351M8.1
|
Uncharacterized protein |
| chr13_+_103459704 | 0.03 |
ENST00000602836.1
|
BIVM-ERCC5
|
BIVM-ERCC5 readthrough |
| chr9_-_79267432 | 0.03 |
ENST00000424866.1
|
PRUNE2
|
prune homolog 2 (Drosophila) |
| chr1_-_154600421 | 0.03 |
ENST00000368471.3
ENST00000292205.5 |
ADAR
|
adenosine deaminase, RNA-specific |
| chr19_+_56280507 | 0.03 |
ENST00000341750.4
|
RFPL4AL1
|
ret finger protein-like 4A-like 1 |
| chr15_+_83209620 | 0.03 |
ENST00000568285.1
|
RP11-379H8.1
|
Uncharacterized protein |
| chr7_-_36764142 | 0.03 |
ENST00000258749.5
ENST00000535891.1 |
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr6_-_169582835 | 0.03 |
ENST00000564830.1
|
RP11-417E7.2
|
RP11-417E7.2 |
| chr12_-_15038779 | 0.03 |
ENST00000228938.5
ENST00000539261.1 |
MGP
|
matrix Gla protein |
| chr2_-_198540719 | 0.03 |
ENST00000295049.4
|
RFTN2
|
raftlin family member 2 |
| chr14_+_101123580 | 0.02 |
ENST00000556697.1
ENST00000360899.2 ENST00000553623.1 |
LINC00523
|
long intergenic non-protein coding RNA 523 |
| chr16_-_20566616 | 0.02 |
ENST00000569163.1
|
ACSM2B
|
acyl-CoA synthetase medium-chain family member 2B |
| chr3_-_49466686 | 0.02 |
ENST00000273598.3
ENST00000436744.2 |
NICN1
|
nicolin 1 |
| chrX_-_101694853 | 0.02 |
ENST00000372749.1
|
NXF2B
|
nuclear RNA export factor 2B |
| chr16_-_71518504 | 0.02 |
ENST00000565100.2
|
ZNF19
|
zinc finger protein 19 |
| chr2_+_1417228 | 0.02 |
ENST00000382269.3
ENST00000337415.3 ENST00000345913.4 ENST00000346956.3 ENST00000349624.3 ENST00000539820.1 ENST00000329066.4 ENST00000382201.3 |
TPO
|
thyroid peroxidase |
| chr9_-_4666421 | 0.02 |
ENST00000381895.5
|
SPATA6L
|
spermatogenesis associated 6-like |
| chr9_-_104198042 | 0.02 |
ENST00000374855.4
|
ALDOB
|
aldolase B, fructose-bisphosphate |
| chr6_-_33864684 | 0.02 |
ENST00000506222.2
ENST00000533304.1 |
LINC01016
|
long intergenic non-protein coding RNA 1016 |
| chr6_+_42141029 | 0.02 |
ENST00000372958.1
|
GUCA1A
|
guanylate cyclase activator 1A (retina) |
| chr22_+_40322595 | 0.02 |
ENST00000420971.1
ENST00000544756.1 |
GRAP2
|
GRB2-related adaptor protein 2 |
| chr4_-_88450595 | 0.01 |
ENST00000434434.1
|
SPARCL1
|
SPARC-like 1 (hevin) |
| chr7_+_20686946 | 0.01 |
ENST00000443026.2
ENST00000406935.1 |
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr4_-_17513851 | 0.01 |
ENST00000281243.5
|
QDPR
|
quinoid dihydropteridine reductase |
| chr8_-_7220490 | 0.01 |
ENST00000400078.2
|
ZNF705G
|
zinc finger protein 705G |
| chr22_+_40322623 | 0.01 |
ENST00000399090.2
|
GRAP2
|
GRB2-related adaptor protein 2 |
| chr6_-_31514333 | 0.01 |
ENST00000376151.4
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr3_-_133648656 | 0.01 |
ENST00000408895.2
|
C3orf36
|
chromosome 3 open reading frame 36 |
| chr12_+_12223867 | 0.01 |
ENST00000308721.5
|
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chr4_-_88450612 | 0.01 |
ENST00000418378.1
ENST00000282470.6 |
SPARCL1
|
SPARC-like 1 (hevin) |
| chr9_+_71357171 | 0.01 |
ENST00000440050.1
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta |
| chr7_-_137028498 | 0.01 |
ENST00000393083.2
|
PTN
|
pleiotrophin |
| chr11_+_94038759 | 0.01 |
ENST00000440961.2
ENST00000328458.4 |
FOLR4
|
folate receptor 4, delta (putative) |
| chr5_+_122181128 | 0.01 |
ENST00000261369.4
|
SNX24
|
sorting nexin 24 |
| chr12_-_11139511 | 0.01 |
ENST00000506868.1
|
TAS2R50
|
taste receptor, type 2, member 50 |
| chr11_-_119249805 | 0.01 |
ENST00000527843.1
|
USP2
|
ubiquitin specific peptidase 2 |
| chr3_-_139199565 | 0.01 |
ENST00000511956.1
|
RBP2
|
retinol binding protein 2, cellular |
| chr16_+_72088376 | 0.01 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr7_-_137028534 | 0.01 |
ENST00000348225.2
|
PTN
|
pleiotrophin |
| chr2_+_234686976 | 0.01 |
ENST00000389758.3
|
MROH2A
|
maestro heat-like repeat family member 2A |
| chr4_-_120243545 | 0.01 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr11_-_56310757 | 0.01 |
ENST00000528616.2
|
OR5M11
|
olfactory receptor, family 5, subfamily M, member 11 |
| chr8_-_16043780 | 0.01 |
ENST00000445506.2
|
MSR1
|
macrophage scavenger receptor 1 |
| chr16_-_89119355 | 0.01 |
ENST00000537498.1
|
CTD-2555A7.2
|
CTD-2555A7.2 |
| chr2_-_166060552 | 0.01 |
ENST00000283254.7
ENST00000453007.1 |
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr8_+_86851932 | 0.00 |
ENST00000517368.1
|
CTA-392E5.1
|
CTA-392E5.1 |
| chr6_+_36839616 | 0.00 |
ENST00000359359.2
ENST00000510325.2 |
C6orf89
|
chromosome 6 open reading frame 89 |
| chr16_+_446713 | 0.00 |
ENST00000397722.1
ENST00000454619.1 |
NME4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr19_-_2085323 | 0.00 |
ENST00000591638.1
|
MOB3A
|
MOB kinase activator 3A |
| chr9_+_42717234 | 0.00 |
ENST00000377590.1
|
FOXD4L2
|
forkhead box D4-like 2 |
| chr1_-_91813006 | 0.00 |
ENST00000430465.1
|
HFM1
|
HFM1, ATP-dependent DNA helicase homolog (S. cerevisiae) |
Network of associatons between targets according to the STRING database.
First level regulatory network of AIRE
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.5 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.4 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.1 | GO:0030451 | regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) |
| 0.0 | 0.3 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.3 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0090095 | regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.0 | 0.1 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.0 | 0.4 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.2 | GO:0032485 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.2 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.5 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 1.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 1.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.8 | GO:0044364 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.1 | GO:0086036 | regulation of cardiac muscle cell membrane potential(GO:0086036) |
| 0.0 | 0.9 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.0 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.0 | 0.2 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.0 | GO:0030070 | insulin processing(GO:0030070) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.5 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0036338 | viral envelope(GO:0019031) viral membrane(GO:0036338) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:0036457 | keratohyalin granule(GO:0036457) |
| 0.0 | 0.2 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 1.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 1.5 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 1.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.4 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0004877 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.2 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.5 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 1.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.0 | 0.6 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |