Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
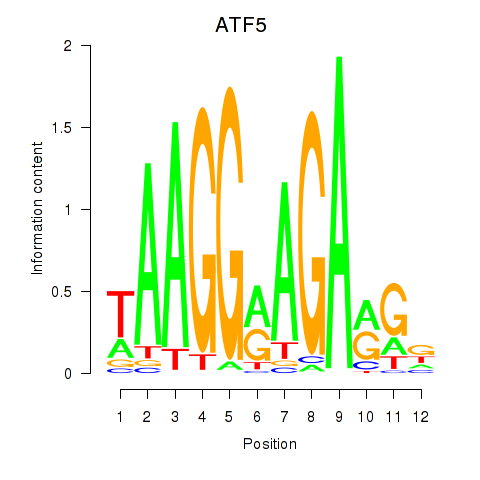
Results for ATF5
Z-value: 1.03
Transcription factors associated with ATF5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ATF5
|
ENSG00000169136.4 | activating transcription factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ATF5 | hg19_v2_chr19_+_50432400_50432479 | -0.38 | 9.4e-02 | Click! |
Activity profile of ATF5 motif
Sorted Z-values of ATF5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_36988882 | 5.45 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr1_+_209859510 | 4.20 |
ENST00000367028.2
ENST00000261465.1 |
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr2_-_113542063 | 4.19 |
ENST00000263339.3
|
IL1A
|
interleukin 1, alpha |
| chr1_-_17304771 | 4.05 |
ENST00000375534.3
|
MFAP2
|
microfibrillar-associated protein 2 |
| chr21_-_28215332 | 3.94 |
ENST00000517777.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr7_-_3083573 | 3.76 |
ENST00000396946.4
|
CARD11
|
caspase recruitment domain family, member 11 |
| chr7_-_41740181 | 3.32 |
ENST00000442711.1
|
INHBA
|
inhibin, beta A |
| chr6_+_43738444 | 2.97 |
ENST00000324450.6
ENST00000417285.2 ENST00000413642.3 ENST00000372055.4 ENST00000482630.2 ENST00000425836.2 ENST00000372064.4 ENST00000372077.4 ENST00000519767.1 |
VEGFA
|
vascular endothelial growth factor A |
| chr5_-_39219641 | 2.91 |
ENST00000509072.1
ENST00000504542.1 ENST00000505428.1 ENST00000506557.1 |
FYB
|
FYN binding protein |
| chr7_-_127672146 | 2.87 |
ENST00000476782.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr5_+_156887027 | 2.78 |
ENST00000435489.2
ENST00000311946.7 |
NIPAL4
|
NIPA-like domain containing 4 |
| chr1_-_46598371 | 2.74 |
ENST00000372006.1
ENST00000425892.1 ENST00000420542.1 ENST00000354242.4 ENST00000340332.6 |
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr7_-_3083472 | 2.65 |
ENST00000356408.3
|
CARD11
|
caspase recruitment domain family, member 11 |
| chr16_-_65155833 | 2.52 |
ENST00000566827.1
ENST00000394156.3 ENST00000562998.1 |
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr1_-_27286897 | 2.48 |
ENST00000320567.5
|
C1orf172
|
chromosome 1 open reading frame 172 |
| chr10_+_111765562 | 2.34 |
ENST00000360162.3
|
ADD3
|
adducin 3 (gamma) |
| chr14_-_65569244 | 2.31 |
ENST00000557277.1
ENST00000556892.1 |
MAX
|
MYC associated factor X |
| chr5_+_50678921 | 2.28 |
ENST00000230658.7
|
ISL1
|
ISL LIM homeobox 1 |
| chr17_-_5522731 | 2.20 |
ENST00000576905.1
|
NLRP1
|
NLR family, pyrin domain containing 1 |
| chr1_+_16083154 | 2.08 |
ENST00000375771.1
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr16_-_65155979 | 2.05 |
ENST00000562325.1
ENST00000268603.4 |
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chrX_+_131157609 | 2.02 |
ENST00000496850.1
|
MST4
|
Serine/threonine-protein kinase MST4 |
| chr1_-_202776392 | 1.93 |
ENST00000235790.4
|
KDM5B
|
lysine (K)-specific demethylase 5B |
| chr5_+_138629628 | 1.91 |
ENST00000508689.1
ENST00000514528.1 |
MATR3
|
matrin 3 |
| chr9_+_109625378 | 1.85 |
ENST00000277225.5
ENST00000457913.1 ENST00000472574.1 |
ZNF462
|
zinc finger protein 462 |
| chr5_+_138629337 | 1.77 |
ENST00000394805.3
ENST00000512876.1 ENST00000513678.1 |
MATR3
|
matrin 3 |
| chr4_+_2800722 | 1.76 |
ENST00000508385.1
|
SH3BP2
|
SH3-domain binding protein 2 |
| chr11_-_117102768 | 1.73 |
ENST00000532301.1
|
PCSK7
|
proprotein convertase subtilisin/kexin type 7 |
| chr11_+_58938903 | 1.65 |
ENST00000532982.1
|
DTX4
|
deltex homolog 4 (Drosophila) |
| chr4_+_77172847 | 1.60 |
ENST00000515604.1
ENST00000539752.1 ENST00000424749.2 |
FAM47E
FAM47E-STBD1
FAM47E
|
uncharacterized protein LOC100631383 FAM47E-STBD1 readthrough family with sequence similarity 47, member E |
| chr12_-_54778444 | 1.57 |
ENST00000551771.1
|
ZNF385A
|
zinc finger protein 385A |
| chr15_+_78556428 | 1.55 |
ENST00000394855.3
ENST00000489435.2 |
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4 |
| chr12_-_54778244 | 1.53 |
ENST00000549937.1
|
ZNF385A
|
zinc finger protein 385A |
| chr5_+_138629389 | 1.51 |
ENST00000504045.1
ENST00000504311.1 ENST00000502499.1 |
MATR3
|
matrin 3 |
| chr3_-_99569821 | 1.50 |
ENST00000487087.1
|
FILIP1L
|
filamin A interacting protein 1-like |
| chr3_-_69129501 | 1.48 |
ENST00000540295.1
ENST00000415609.2 ENST00000361055.4 ENST00000349511.4 |
UBA3
|
ubiquitin-like modifier activating enzyme 3 |
| chr2_-_214016314 | 1.48 |
ENST00000434687.1
ENST00000374319.4 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr11_-_129062093 | 1.46 |
ENST00000310343.9
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr6_-_100016678 | 1.43 |
ENST00000523799.1
ENST00000520429.1 |
CCNC
|
cyclin C |
| chr22_-_36236623 | 1.40 |
ENST00000405409.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr17_+_38334242 | 1.39 |
ENST00000436615.3
|
RAPGEFL1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr7_-_127671674 | 1.38 |
ENST00000478726.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr5_-_39219705 | 1.33 |
ENST00000351578.6
|
FYB
|
FYN binding protein |
| chr16_-_58663720 | 1.32 |
ENST00000564557.1
ENST00000569240.1 ENST00000441024.2 ENST00000569020.1 ENST00000317147.5 |
CNOT1
|
CCR4-NOT transcription complex, subunit 1 |
| chr12_-_57030096 | 1.30 |
ENST00000549506.1
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chr7_-_102232891 | 1.28 |
ENST00000514917.2
|
RP11-514P8.7
|
RP11-514P8.7 |
| chr7_+_128399002 | 1.27 |
ENST00000493278.1
|
CALU
|
calumenin |
| chr19_+_41509851 | 1.27 |
ENST00000593831.1
ENST00000330446.5 |
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6 |
| chr6_+_87865262 | 1.26 |
ENST00000369577.3
ENST00000518845.1 ENST00000339907.4 ENST00000496806.2 |
ZNF292
|
zinc finger protein 292 |
| chr1_-_54872059 | 1.25 |
ENST00000371320.3
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr17_-_79817091 | 1.25 |
ENST00000570907.1
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr7_+_132937820 | 1.23 |
ENST00000393161.2
ENST00000253861.4 |
EXOC4
|
exocyst complex component 4 |
| chr1_+_101361782 | 1.21 |
ENST00000357650.4
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr5_+_82767583 | 1.20 |
ENST00000512590.2
ENST00000513960.1 ENST00000513984.1 ENST00000502527.2 |
VCAN
|
versican |
| chr16_+_9185450 | 1.19 |
ENST00000327827.7
|
C16orf72
|
chromosome 16 open reading frame 72 |
| chr4_-_74124502 | 1.17 |
ENST00000358602.4
ENST00000330838.6 ENST00000561029.1 |
ANKRD17
|
ankyrin repeat domain 17 |
| chr3_-_183145873 | 1.17 |
ENST00000447025.2
ENST00000414362.2 ENST00000328913.3 |
MCF2L2
|
MCF.2 cell line derived transforming sequence-like 2 |
| chr14_+_32546145 | 1.17 |
ENST00000556611.1
ENST00000539826.2 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr5_+_82767487 | 1.15 |
ENST00000343200.5
ENST00000342785.4 |
VCAN
|
versican |
| chr15_+_63481668 | 1.15 |
ENST00000321437.4
ENST00000559006.1 ENST00000448330.2 |
RAB8B
|
RAB8B, member RAS oncogene family |
| chr5_+_82767284 | 1.13 |
ENST00000265077.3
|
VCAN
|
versican |
| chr14_-_65569057 | 1.11 |
ENST00000555419.1
ENST00000341653.2 |
MAX
|
MYC associated factor X |
| chr12_+_56915776 | 1.09 |
ENST00000550726.1
ENST00000542360.1 |
RBMS2
|
RNA binding motif, single stranded interacting protein 2 |
| chr8_-_131028869 | 1.09 |
ENST00000518283.1
ENST00000519110.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr6_+_44095347 | 1.07 |
ENST00000323267.6
|
TMEM63B
|
transmembrane protein 63B |
| chr1_-_67896069 | 1.07 |
ENST00000370995.2
ENST00000361219.6 |
SERBP1
|
SERPINE1 mRNA binding protein 1 |
| chr12_-_54778471 | 1.07 |
ENST00000550120.1
ENST00000394313.2 ENST00000547210.1 |
ZNF385A
|
zinc finger protein 385A |
| chr14_-_65569186 | 1.05 |
ENST00000555932.1
ENST00000358664.4 ENST00000284165.6 ENST00000358402.4 ENST00000246163.2 ENST00000556979.1 ENST00000555667.1 ENST00000557746.1 ENST00000556443.1 |
MAX
|
MYC associated factor X |
| chr6_+_44095263 | 1.04 |
ENST00000532634.1
|
TMEM63B
|
transmembrane protein 63B |
| chr16_+_72127806 | 1.03 |
ENST00000566489.1
|
DHX38
|
DEAH (Asp-Glu-Ala-His) box polypeptide 38 |
| chr3_-_187454281 | 1.02 |
ENST00000232014.4
|
BCL6
|
B-cell CLL/lymphoma 6 |
| chr2_+_191334212 | 1.01 |
ENST00000444317.1
ENST00000535751.1 |
MFSD6
|
major facilitator superfamily domain containing 6 |
| chr14_+_70918874 | 1.01 |
ENST00000603540.1
|
ADAM21
|
ADAM metallopeptidase domain 21 |
| chr1_+_16083098 | 0.98 |
ENST00000496928.2
ENST00000508310.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr18_-_812517 | 0.98 |
ENST00000584307.1
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr22_-_36357671 | 0.97 |
ENST00000408983.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr5_-_148442584 | 0.96 |
ENST00000394358.2
ENST00000512049.1 |
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr12_+_32655048 | 0.96 |
ENST00000427716.2
ENST00000266482.3 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr14_+_96968802 | 0.96 |
ENST00000556619.1
ENST00000392990.2 |
PAPOLA
|
poly(A) polymerase alpha |
| chr18_-_812231 | 0.92 |
ENST00000314574.4
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr19_+_42773371 | 0.92 |
ENST00000571942.2
|
CIC
|
capicua transcriptional repressor |
| chr6_-_100016527 | 0.90 |
ENST00000523985.1
ENST00000518714.1 ENST00000520371.1 |
CCNC
|
cyclin C |
| chr11_+_27076764 | 0.88 |
ENST00000525090.1
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr12_+_56915713 | 0.85 |
ENST00000262031.5
ENST00000552247.2 |
RBMS2
|
RNA binding motif, single stranded interacting protein 2 |
| chr2_-_197036289 | 0.85 |
ENST00000263955.4
|
STK17B
|
serine/threonine kinase 17b |
| chr12_+_32655110 | 0.85 |
ENST00000546442.1
ENST00000583694.1 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr20_+_58179582 | 0.84 |
ENST00000371015.1
ENST00000395639.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr1_-_27930102 | 0.83 |
ENST00000247087.5
ENST00000374011.2 |
AHDC1
|
AT hook, DNA binding motif, containing 1 |
| chr13_-_36944307 | 0.82 |
ENST00000355182.4
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome) |
| chr7_+_44040488 | 0.81 |
ENST00000258704.3
|
SPDYE1
|
speedy/RINGO cell cycle regulator family member E1 |
| chr3_+_190105909 | 0.81 |
ENST00000456423.1
|
CLDN16
|
claudin 16 |
| chr1_-_67896009 | 0.78 |
ENST00000370990.5
|
SERBP1
|
SERPINE1 mRNA binding protein 1 |
| chr8_+_79428539 | 0.77 |
ENST00000352966.5
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
| chr20_-_52612705 | 0.76 |
ENST00000434986.2
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr2_+_234601512 | 0.75 |
ENST00000305139.6
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr6_-_39902160 | 0.75 |
ENST00000340692.5
|
MOCS1
|
molybdenum cofactor synthesis 1 |
| chr5_+_50679506 | 0.74 |
ENST00000511384.1
|
ISL1
|
ISL LIM homeobox 1 |
| chr5_-_148442687 | 0.72 |
ENST00000515425.1
|
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr1_+_101361626 | 0.71 |
ENST00000370112.4
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr12_+_32654965 | 0.70 |
ENST00000472289.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr17_+_55055466 | 0.70 |
ENST00000262288.3
ENST00000572710.1 ENST00000575395.1 |
SCPEP1
|
serine carboxypeptidase 1 |
| chr2_-_128642434 | 0.69 |
ENST00000393001.1
|
AMMECR1L
|
AMMECR1-like |
| chr12_-_52967600 | 0.69 |
ENST00000549343.1
ENST00000305620.2 |
KRT74
|
keratin 74 |
| chr19_-_15090488 | 0.67 |
ENST00000594383.1
ENST00000598504.1 ENST00000597262.1 |
SLC1A6
|
solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 |
| chr12_-_72057638 | 0.67 |
ENST00000552037.1
ENST00000378743.3 |
ZFC3H1
|
zinc finger, C3H1-type containing |
| chr16_+_72127461 | 0.64 |
ENST00000268482.3
ENST00000566794.1 |
DHX38
|
DEAH (Asp-Glu-Ala-His) box polypeptide 38 |
| chr2_+_217498105 | 0.63 |
ENST00000233809.4
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa |
| chr6_+_143999185 | 0.63 |
ENST00000542769.1
ENST00000397980.3 |
PHACTR2
|
phosphatase and actin regulator 2 |
| chr3_+_38206975 | 0.63 |
ENST00000446845.1
ENST00000311806.3 |
OXSR1
|
oxidative stress responsive 1 |
| chr14_+_32547434 | 0.63 |
ENST00000556191.1
ENST00000554090.1 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr1_-_22109484 | 0.63 |
ENST00000529637.1
|
USP48
|
ubiquitin specific peptidase 48 |
| chr1_-_151319318 | 0.61 |
ENST00000436271.1
ENST00000450506.1 ENST00000422595.1 |
RFX5
|
regulatory factor X, 5 (influences HLA class II expression) |
| chr11_+_124735282 | 0.59 |
ENST00000397801.1
|
ROBO3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr1_+_160321120 | 0.58 |
ENST00000424754.1
|
NCSTN
|
nicastrin |
| chr12_-_57030115 | 0.58 |
ENST00000379441.3
ENST00000179765.5 ENST00000551812.1 |
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chr12_-_49351303 | 0.57 |
ENST00000256682.4
|
ARF3
|
ADP-ribosylation factor 3 |
| chr3_+_193853927 | 0.56 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr6_+_76599809 | 0.56 |
ENST00000430435.1
|
MYO6
|
myosin VI |
| chr10_+_103113840 | 0.55 |
ENST00000393441.4
ENST00000408038.2 |
BTRC
|
beta-transducin repeat containing E3 ubiquitin protein ligase |
| chr9_+_127023704 | 0.55 |
ENST00000373596.1
ENST00000425237.1 |
NEK6
|
NIMA-related kinase 6 |
| chr18_-_53089723 | 0.53 |
ENST00000561992.1
ENST00000562512.2 |
TCF4
|
transcription factor 4 |
| chr2_-_128784846 | 0.53 |
ENST00000259235.3
ENST00000357702.5 ENST00000424298.1 |
SAP130
|
Sin3A-associated protein, 130kDa |
| chr19_-_45873642 | 0.52 |
ENST00000485403.2
ENST00000586856.1 ENST00000586131.1 ENST00000391940.4 ENST00000221481.6 ENST00000391944.3 ENST00000391945.4 |
ERCC2
|
excision repair cross-complementing rodent repair deficiency, complementation group 2 |
| chrX_+_123095546 | 0.52 |
ENST00000371157.3
ENST00000371145.3 ENST00000371144.3 |
STAG2
|
stromal antigen 2 |
| chr1_+_16083123 | 0.52 |
ENST00000510393.1
ENST00000430076.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr1_-_36023251 | 0.51 |
ENST00000426982.2
|
KIAA0319L
|
KIAA0319-like |
| chr7_+_143771275 | 0.49 |
ENST00000408898.2
|
OR2A25
|
olfactory receptor, family 2, subfamily A, member 25 |
| chr11_-_108408895 | 0.49 |
ENST00000443411.1
ENST00000533052.1 |
EXPH5
|
exophilin 5 |
| chr5_+_138629417 | 0.47 |
ENST00000510056.1
ENST00000511249.1 ENST00000503811.1 ENST00000511378.1 |
MATR3
|
matrin 3 |
| chr5_+_34757309 | 0.47 |
ENST00000397449.1
|
RAI14
|
retinoic acid induced 14 |
| chr17_+_33914424 | 0.46 |
ENST00000590432.1
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr11_+_28724129 | 0.46 |
ENST00000513853.1
|
RP11-115J23.1
|
RP11-115J23.1 |
| chr1_-_22109682 | 0.46 |
ENST00000400301.1
ENST00000532737.1 |
USP48
|
ubiquitin specific peptidase 48 |
| chr6_-_100016492 | 0.45 |
ENST00000369217.4
ENST00000369220.4 ENST00000482541.2 |
CCNC
|
cyclin C |
| chr17_+_38337491 | 0.45 |
ENST00000538981.1
|
RAPGEFL1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr12_-_12491608 | 0.42 |
ENST00000545735.1
|
MANSC1
|
MANSC domain containing 1 |
| chr1_-_205053645 | 0.42 |
ENST00000367167.3
|
TMEM81
|
transmembrane protein 81 |
| chr1_-_151319283 | 0.41 |
ENST00000392746.3
|
RFX5
|
regulatory factor X, 5 (influences HLA class II expression) |
| chr9_+_2621950 | 0.39 |
ENST00000382096.1
|
VLDLR
|
very low density lipoprotein receptor |
| chr12_-_72057571 | 0.39 |
ENST00000548100.1
|
ZFC3H1
|
zinc finger, C3H1-type containing |
| chr5_+_137203465 | 0.38 |
ENST00000239926.4
|
MYOT
|
myotilin |
| chr3_+_19988885 | 0.37 |
ENST00000422242.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr1_+_61869748 | 0.37 |
ENST00000357977.5
|
NFIA
|
nuclear factor I/A |
| chr1_+_36023370 | 0.36 |
ENST00000356090.4
ENST00000373243.2 |
NCDN
|
neurochondrin |
| chr3_-_23958402 | 0.35 |
ENST00000415901.2
ENST00000416026.2 ENST00000412028.1 ENST00000388759.3 ENST00000437230.1 |
NKIRAS1
|
NFKB inhibitor interacting Ras-like 1 |
| chr3_-_182698381 | 0.35 |
ENST00000292782.4
|
DCUN1D1
|
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chr3_-_47950745 | 0.35 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr6_+_4773205 | 0.35 |
ENST00000440139.1
|
CDYL
|
chromodomain protein, Y-like |
| chrX_+_47050236 | 0.34 |
ENST00000377351.4
|
UBA1
|
ubiquitin-like modifier activating enzyme 1 |
| chr1_+_151739131 | 0.34 |
ENST00000400999.1
|
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr9_-_116065551 | 0.33 |
ENST00000297894.5
|
RNF183
|
ring finger protein 183 |
| chr14_+_100111447 | 0.33 |
ENST00000330710.5
ENST00000357223.2 |
HHIPL1
|
HHIP-like 1 |
| chr1_+_36023035 | 0.33 |
ENST00000373253.3
|
NCDN
|
neurochondrin |
| chr9_-_23825956 | 0.33 |
ENST00000397312.2
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr1_-_156460391 | 0.32 |
ENST00000360595.3
|
MEF2D
|
myocyte enhancer factor 2D |
| chr11_-_7904464 | 0.32 |
ENST00000502624.2
ENST00000527565.1 |
RP11-35J10.5
|
RP11-35J10.5 |
| chr3_+_19988566 | 0.29 |
ENST00000273047.4
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr8_+_100025476 | 0.29 |
ENST00000355155.1
ENST00000357162.2 ENST00000358544.2 ENST00000395996.1 ENST00000441350.2 |
VPS13B
|
vacuolar protein sorting 13 homolog B (yeast) |
| chr11_-_33891362 | 0.28 |
ENST00000395833.3
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr3_-_23958506 | 0.26 |
ENST00000425478.2
|
NKIRAS1
|
NFKB inhibitor interacting Ras-like 1 |
| chr4_-_84376953 | 0.26 |
ENST00000510985.1
ENST00000295488.3 |
HELQ
|
helicase, POLQ-like |
| chr1_+_206579736 | 0.26 |
ENST00000439126.1
|
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr17_+_44790515 | 0.26 |
ENST00000576346.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr9_+_19230433 | 0.25 |
ENST00000434457.2
ENST00000602925.1 |
DENND4C
|
DENN/MADD domain containing 4C |
| chr17_+_38334501 | 0.25 |
ENST00000541245.1
|
RAPGEFL1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr17_-_73149921 | 0.24 |
ENST00000481647.1
ENST00000470924.1 |
HN1
|
hematological and neurological expressed 1 |
| chr15_+_77287426 | 0.23 |
ENST00000558012.1
ENST00000267939.5 ENST00000379595.3 |
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1 |
| chrX_-_19983260 | 0.23 |
ENST00000340625.3
|
CXorf23
|
chromosome X open reading frame 23 |
| chr12_+_125811162 | 0.23 |
ENST00000299308.3
|
TMEM132B
|
transmembrane protein 132B |
| chr5_+_137203541 | 0.22 |
ENST00000421631.2
|
MYOT
|
myotilin |
| chr12_-_25101920 | 0.22 |
ENST00000539780.1
ENST00000546285.1 ENST00000342945.5 |
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr4_-_68620053 | 0.22 |
ENST00000420975.2
ENST00000226413.4 |
GNRHR
|
gonadotropin-releasing hormone receptor |
| chr17_+_44668387 | 0.21 |
ENST00000576040.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr2_+_88047606 | 0.21 |
ENST00000359481.4
|
PLGLB2
|
plasminogen-like B2 |
| chr6_+_114178512 | 0.20 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr6_+_31865552 | 0.19 |
ENST00000469372.1
ENST00000497706.1 |
C2
|
complement component 2 |
| chr16_+_28943260 | 0.19 |
ENST00000538922.1
ENST00000324662.3 ENST00000567541.1 |
CD19
|
CD19 molecule |
| chr7_-_115670792 | 0.18 |
ENST00000265440.7
ENST00000393485.1 |
TFEC
|
transcription factor EC |
| chrX_+_153409678 | 0.18 |
ENST00000369951.4
|
OPN1LW
|
opsin 1 (cone pigments), long-wave-sensitive |
| chr5_+_137203557 | 0.17 |
ENST00000515645.1
|
MYOT
|
myotilin |
| chr9_-_23826298 | 0.17 |
ENST00000380117.1
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr8_-_623547 | 0.17 |
ENST00000522893.1
|
ERICH1
|
glutamate-rich 1 |
| chr17_+_33914460 | 0.16 |
ENST00000537622.2
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr5_+_125967372 | 0.14 |
ENST00000357147.3
|
C5orf48
|
chromosome 5 open reading frame 48 |
| chr8_+_37553261 | 0.14 |
ENST00000331569.4
|
ZNF703
|
zinc finger protein 703 |
| chr8_-_13134045 | 0.13 |
ENST00000512044.2
|
DLC1
|
deleted in liver cancer 1 |
| chr20_-_52612468 | 0.13 |
ENST00000422805.1
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr18_-_53089538 | 0.13 |
ENST00000566777.1
|
TCF4
|
transcription factor 4 |
| chr19_+_15160130 | 0.13 |
ENST00000427043.3
|
CASP14
|
caspase 14, apoptosis-related cysteine peptidase |
| chrX_-_122866874 | 0.13 |
ENST00000245838.8
ENST00000355725.4 |
THOC2
|
THO complex 2 |
| chr19_-_38916839 | 0.12 |
ENST00000433821.2
ENST00000426920.2 ENST00000587753.1 ENST00000454404.2 ENST00000293062.9 |
RASGRP4
|
RAS guanyl releasing protein 4 |
| chr12_-_48597170 | 0.12 |
ENST00000310248.2
|
OR10AD1
|
olfactory receptor, family 10, subfamily AD, member 1 |
| chr13_-_36871886 | 0.11 |
ENST00000491049.2
ENST00000503173.1 ENST00000239860.6 ENST00000379862.2 ENST00000239859.7 ENST00000379864.2 ENST00000510088.1 ENST00000554962.1 ENST00000511166.1 |
CCDC169
SOHLH2
CCDC169-SOHLH2
|
coiled-coil domain containing 169 spermatogenesis and oogenesis specific basic helix-loop-helix 2 CCDC169-SOHLH2 readthrough |
| chr8_+_107738240 | 0.11 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr7_-_115670804 | 0.11 |
ENST00000320239.7
|
TFEC
|
transcription factor EC |
| chr10_-_104179682 | 0.11 |
ENST00000406432.1
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr2_-_208994548 | 0.11 |
ENST00000282141.3
|
CRYGC
|
crystallin, gamma C |
| chr11_+_18417948 | 0.11 |
ENST00000542179.1
|
LDHA
|
lactate dehydrogenase A |
| chr22_-_36236265 | 0.10 |
ENST00000414461.2
ENST00000416721.2 ENST00000449924.2 ENST00000262829.7 ENST00000397305.3 |
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr11_-_132813566 | 0.10 |
ENST00000331898.7
|
OPCML
|
opioid binding protein/cell adhesion molecule-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of ATF5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 1.0 | 3.0 | GO:1903570 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.8 | 3.0 | GO:0048936 | visceral motor neuron differentiation(GO:0021524) peripheral nervous system neuron axonogenesis(GO:0048936) cardiac cell fate determination(GO:0060913) positive regulation of granulocyte colony-stimulating factor production(GO:0071657) positive regulation of macrophage colony-stimulating factor production(GO:1901258) |
| 0.7 | 2.2 | GO:1904784 | NLRP1 inflammasome complex assembly(GO:1904784) |
| 0.6 | 4.2 | GO:1902162 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.6 | 10.6 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.5 | 1.5 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.5 | 1.9 | GO:0034721 | histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) |
| 0.5 | 5.4 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.5 | 4.1 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.4 | 1.2 | GO:0097065 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.3 | 1.0 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.2 | 2.5 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.2 | 2.8 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.2 | 4.3 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.2 | 0.6 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.2 | 0.8 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.2 | 1.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.2 | 0.6 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) negative regulation of auditory receptor cell differentiation(GO:0045608) regulation of timing of neuron differentiation(GO:0060164) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.2 | 3.7 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.2 | 4.2 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.2 | 0.5 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.2 | 2.8 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 1.1 | GO:0051461 | positive regulation of corticotropin secretion(GO:0051461) |
| 0.2 | 4.2 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.1 | 1.9 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 3.5 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 1.3 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 1.2 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 | 1.7 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 | 1.3 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.1 | 0.7 | GO:0070778 | L-aspartate transport(GO:0070778) L-aspartate transmembrane transport(GO:0089712) |
| 0.1 | 4.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 1.7 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 0.8 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.1 | 0.7 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 2.7 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.1 | 1.9 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.1 | 1.2 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.1 | 0.7 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.1 | 0.3 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.9 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 1.6 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 | 0.3 | GO:0021816 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.1 | 0.8 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.4 | GO:2000568 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.1 | 0.9 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 1.9 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.1 | 0.6 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 0.5 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.3 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0002372 | myeloid dendritic cell cytokine production(GO:0002372) |
| 0.0 | 0.2 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.2 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 1.7 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 1.8 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.6 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 1.0 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.7 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.0 | 0.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 2.3 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 2.3 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 4.2 | GO:0009267 | cellular response to starvation(GO:0009267) |
| 0.0 | 0.6 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.7 | GO:0033598 | mammary gland epithelial cell proliferation(GO:0033598) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.6 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.3 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.2 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.3 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.1 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.3 | 2.2 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.3 | 1.2 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.3 | 1.9 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.2 | 0.7 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.2 | 6.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.2 | 4.1 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 1.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 1.1 | GO:0051286 | cell tip(GO:0051286) |
| 0.1 | 1.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.5 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 1.3 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 4.5 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 2.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 2.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.6 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 4.8 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.7 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 3.1 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 3.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 3.5 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 3.0 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 5.2 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 2.5 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 2.3 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.3 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.5 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 7.8 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.5 | 3.0 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.5 | 1.5 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.5 | 1.9 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.5 | 2.9 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.3 | 6.5 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.3 | 6.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 0.9 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.2 | 0.6 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.2 | 2.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.2 | 4.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 3.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 2.2 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.1 | 4.9 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 3.6 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.7 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 1.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.1 | 3.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.7 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 1.0 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.2 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.1 | 3.9 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.1 | 0.3 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.4 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.1 | 1.3 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 2.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.5 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 1.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.6 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 3.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 1.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 5.9 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.2 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.7 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 4.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.6 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.0 | 0.7 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.8 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.7 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 3.2 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 3.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.8 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 1.5 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.8 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 2.1 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.7 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 2.6 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 3.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 12.4 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 4.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 2.7 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 2.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 3.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 1.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 4.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 5.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.9 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 4.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.2 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.6 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 4.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 3.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 3.0 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 3.0 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 3.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 6.4 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 4.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 4.6 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 3.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 4.2 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 3.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 4.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 1.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 2.5 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 3.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 1.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 2.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 4.5 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 2.2 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 2.0 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.3 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.7 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 2.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.8 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.6 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 2.0 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.6 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |