Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
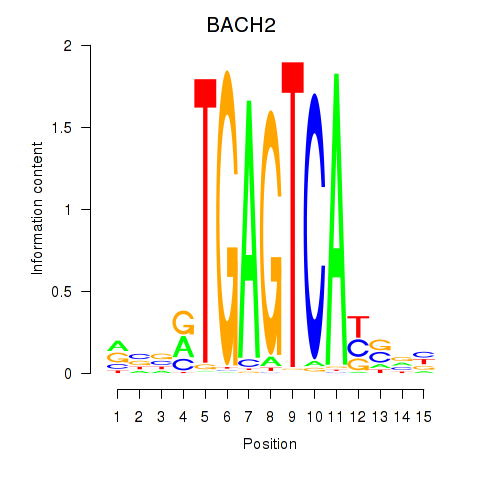
Results for BACH2
Z-value: 0.64
Transcription factors associated with BACH2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BACH2
|
ENSG00000112182.10 | BTB domain and CNC homolog 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BACH2 | hg19_v2_chr6_-_91006461_91006517 | 0.75 | 1.6e-04 | Click! |
Activity profile of BACH2 motif
Sorted Z-values of BACH2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_39769005 | 6.53 |
ENST00000301653.4
ENST00000593067.1 |
KRT16
|
keratin 16 |
| chr2_+_113875466 | 4.70 |
ENST00000361779.3
ENST00000259206.5 ENST00000354115.2 |
IL1RN
|
interleukin 1 receptor antagonist |
| chr1_+_153003671 | 3.80 |
ENST00000307098.4
|
SPRR1B
|
small proline-rich protein 1B |
| chr1_+_152881014 | 3.52 |
ENST00000368764.3
ENST00000392667.2 |
IVL
|
involucrin |
| chr2_+_208104351 | 3.25 |
ENST00000440326.1
|
AC007879.7
|
AC007879.7 |
| chr3_-_151034734 | 3.21 |
ENST00000260843.4
|
GPR87
|
G protein-coupled receptor 87 |
| chr18_+_21452964 | 3.15 |
ENST00000587184.1
|
LAMA3
|
laminin, alpha 3 |
| chr18_+_21452804 | 3.14 |
ENST00000269217.6
|
LAMA3
|
laminin, alpha 3 |
| chr2_+_208104497 | 3.00 |
ENST00000430494.1
|
AC007879.7
|
AC007879.7 |
| chr11_-_119993979 | 2.64 |
ENST00000524816.3
ENST00000525327.1 |
TRIM29
|
tripartite motif containing 29 |
| chr1_-_156675368 | 2.64 |
ENST00000368222.3
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr18_+_61143994 | 2.41 |
ENST00000382771.4
|
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr20_-_1309809 | 2.19 |
ENST00000360779.3
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr6_+_41604747 | 2.14 |
ENST00000419164.1
ENST00000373051.2 |
MDFI
|
MyoD family inhibitor |
| chr10_+_75668916 | 1.97 |
ENST00000481390.1
|
PLAU
|
plasminogen activator, urokinase |
| chr9_-_114246332 | 1.96 |
ENST00000602978.1
|
KIAA0368
|
KIAA0368 |
| chr20_+_44637526 | 1.96 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr5_+_149877334 | 1.96 |
ENST00000523767.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr6_+_106959718 | 1.92 |
ENST00000369066.3
|
AIM1
|
absent in melanoma 1 |
| chr6_-_113953705 | 1.89 |
ENST00000452675.1
|
RP11-367G18.1
|
RP11-367G18.1 |
| chr16_-_2908155 | 1.88 |
ENST00000571228.1
ENST00000161006.3 |
PRSS22
|
protease, serine, 22 |
| chr3_+_184016986 | 1.88 |
ENST00000417952.1
|
PSMD2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| chr1_+_150480551 | 1.85 |
ENST00000369049.4
ENST00000369047.4 |
ECM1
|
extracellular matrix protein 1 |
| chr11_-_102668879 | 1.80 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr1_+_150480576 | 1.80 |
ENST00000346569.6
|
ECM1
|
extracellular matrix protein 1 |
| chr18_+_61144160 | 1.74 |
ENST00000489441.1
ENST00000424602.1 |
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr19_-_51587502 | 1.70 |
ENST00000156499.2
ENST00000391802.1 |
KLK14
|
kallikrein-related peptidase 14 |
| chr2_+_169926047 | 1.67 |
ENST00000428522.1
ENST00000450153.1 ENST00000421653.1 |
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr6_+_41604620 | 1.66 |
ENST00000432027.1
|
MDFI
|
MyoD family inhibitor |
| chr8_+_144821557 | 1.64 |
ENST00000534398.1
|
FAM83H-AS1
|
FAM83H antisense RNA 1 (head to head) |
| chr19_-_39264072 | 1.60 |
ENST00000599035.1
ENST00000378626.4 |
LGALS7
|
lectin, galactoside-binding, soluble, 7 |
| chr1_+_26606608 | 1.58 |
ENST00000319041.6
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3 |
| chr19_+_39279838 | 1.57 |
ENST00000314980.4
|
LGALS7B
|
lectin, galactoside-binding, soluble, 7B |
| chr11_+_12308447 | 1.52 |
ENST00000256186.2
|
MICALCL
|
MICAL C-terminal like |
| chr12_-_95009837 | 1.51 |
ENST00000551457.1
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr1_+_223889310 | 1.47 |
ENST00000434648.1
|
CAPN2
|
calpain 2, (m/II) large subunit |
| chr8_+_22844995 | 1.47 |
ENST00000524077.1
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr8_+_120220561 | 1.46 |
ENST00000276681.6
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene) |
| chr6_-_35888824 | 1.39 |
ENST00000361690.3
ENST00000512445.1 |
SRPK1
|
SRSF protein kinase 1 |
| chr17_+_37030127 | 1.33 |
ENST00000419929.1
|
LASP1
|
LIM and SH3 protein 1 |
| chr1_+_36621174 | 1.25 |
ENST00000429533.2
|
MAP7D1
|
MAP7 domain containing 1 |
| chr12_-_95010147 | 1.24 |
ENST00000548918.1
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr8_+_123875624 | 1.23 |
ENST00000534247.1
|
ZHX2
|
zinc fingers and homeoboxes 2 |
| chr11_-_102826434 | 1.23 |
ENST00000340273.4
ENST00000260302.3 |
MMP13
|
matrix metallopeptidase 13 (collagenase 3) |
| chr16_+_57662138 | 1.22 |
ENST00000562414.1
ENST00000561969.1 ENST00000562631.1 ENST00000563445.1 ENST00000565338.1 ENST00000567702.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr5_+_150020240 | 1.22 |
ENST00000519664.1
|
SYNPO
|
synaptopodin |
| chr19_-_51523412 | 1.20 |
ENST00000391805.1
ENST00000599077.1 |
KLK10
|
kallikrein-related peptidase 10 |
| chr2_+_220144168 | 1.15 |
ENST00000392087.2
ENST00000442681.1 ENST00000439026.1 |
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr18_+_61442629 | 1.14 |
ENST00000398019.2
ENST00000540675.1 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr11_-_62323702 | 1.12 |
ENST00000530285.1
|
AHNAK
|
AHNAK nucleoprotein |
| chr6_+_74405501 | 1.09 |
ENST00000437994.2
ENST00000422508.2 |
CD109
|
CD109 molecule |
| chr1_+_223889285 | 1.08 |
ENST00000433674.2
|
CAPN2
|
calpain 2, (m/II) large subunit |
| chr8_+_22844913 | 1.08 |
ENST00000519685.1
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr6_+_74405804 | 1.07 |
ENST00000287097.5
|
CD109
|
CD109 molecule |
| chr6_-_35888905 | 1.06 |
ENST00000510290.1
ENST00000423325.2 ENST00000373822.1 |
SRPK1
|
SRSF protein kinase 1 |
| chr19_-_51523275 | 1.05 |
ENST00000309958.3
|
KLK10
|
kallikrein-related peptidase 10 |
| chr19_+_19516561 | 1.01 |
ENST00000457895.2
|
GATAD2A
|
GATA zinc finger domain containing 2A |
| chr2_+_47596287 | 1.00 |
ENST00000263735.4
|
EPCAM
|
epithelial cell adhesion molecule |
| chr19_-_35992780 | 0.99 |
ENST00000593342.1
ENST00000601650.1 ENST00000408915.2 |
DMKN
|
dermokine |
| chr4_-_74088800 | 0.98 |
ENST00000509867.2
|
ANKRD17
|
ankyrin repeat domain 17 |
| chr6_-_56819385 | 0.98 |
ENST00000370754.5
ENST00000449297.2 |
DST
|
dystonin |
| chr7_+_73245193 | 0.94 |
ENST00000340958.2
|
CLDN4
|
claudin 4 |
| chr1_-_6551720 | 0.93 |
ENST00000377728.3
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr11_+_10477733 | 0.92 |
ENST00000528723.1
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr14_+_71165292 | 0.92 |
ENST00000553682.1
|
RP6-65G23.1
|
RP6-65G23.1 |
| chr14_-_75083313 | 0.92 |
ENST00000556652.1
ENST00000555313.1 |
CTD-2207P18.2
|
CTD-2207P18.2 |
| chr2_+_47596634 | 0.91 |
ENST00000419334.1
|
EPCAM
|
epithelial cell adhesion molecule |
| chr6_-_159239257 | 0.91 |
ENST00000337147.7
ENST00000392177.4 |
EZR
|
ezrin |
| chr1_-_52344416 | 0.91 |
ENST00000544028.1
|
NRD1
|
nardilysin (N-arginine dibasic convertase) |
| chr1_+_154377669 | 0.90 |
ENST00000368485.3
ENST00000344086.4 |
IL6R
|
interleukin 6 receptor |
| chr16_+_67571351 | 0.86 |
ENST00000428437.2
ENST00000569253.1 |
FAM65A
|
family with sequence similarity 65, member A |
| chr5_+_138089100 | 0.85 |
ENST00000520339.1
ENST00000355078.5 ENST00000302763.7 ENST00000518910.1 |
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr12_-_48152853 | 0.84 |
ENST00000171000.4
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr7_-_107643567 | 0.84 |
ENST00000393559.1
ENST00000439976.1 ENST00000393560.1 |
LAMB1
|
laminin, beta 1 |
| chr10_+_86088381 | 0.83 |
ENST00000224756.8
ENST00000372088.2 |
CCSER2
|
coiled-coil serine-rich protein 2 |
| chr1_-_27816641 | 0.83 |
ENST00000430629.2
|
WASF2
|
WAS protein family, member 2 |
| chr17_+_48611853 | 0.83 |
ENST00000507709.1
ENST00000515126.1 ENST00000507467.1 |
EPN3
|
epsin 3 |
| chr1_+_26605618 | 0.82 |
ENST00000270792.5
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3 |
| chr3_-_69402828 | 0.81 |
ENST00000460709.1
|
FRMD4B
|
FERM domain containing 4B |
| chr16_+_57662527 | 0.81 |
ENST00000563374.1
ENST00000568234.1 ENST00000565770.1 ENST00000564338.1 ENST00000566164.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr17_+_30771279 | 0.79 |
ENST00000261712.3
ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr2_+_220144052 | 0.79 |
ENST00000425450.1
ENST00000392086.4 ENST00000421532.1 |
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr1_+_156863470 | 0.78 |
ENST00000338302.3
ENST00000455314.1 ENST00000292357.7 |
PEAR1
|
platelet endothelial aggregation receptor 1 |
| chr2_+_231921574 | 0.76 |
ENST00000308696.6
ENST00000373635.4 ENST00000440838.1 ENST00000409643.1 |
PSMD1
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 |
| chr5_-_175843569 | 0.75 |
ENST00000310418.4
ENST00000345807.2 |
CLTB
|
clathrin, light chain B |
| chrX_+_41193407 | 0.75 |
ENST00000457138.2
ENST00000441189.2 |
DDX3X
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr11_-_8739566 | 0.75 |
ENST00000533020.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr4_+_159727222 | 0.74 |
ENST00000512986.1
|
FNIP2
|
folliculin interacting protein 2 |
| chr1_-_108231101 | 0.73 |
ENST00000544443.1
ENST00000415432.2 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr10_+_86088337 | 0.71 |
ENST00000359979.4
|
CCSER2
|
coiled-coil serine-rich protein 2 |
| chr11_-_6341844 | 0.70 |
ENST00000303927.3
|
PRKCDBP
|
protein kinase C, delta binding protein |
| chr7_+_134528635 | 0.69 |
ENST00000445569.2
|
CALD1
|
caldesmon 1 |
| chr6_+_292051 | 0.69 |
ENST00000344450.5
|
DUSP22
|
dual specificity phosphatase 22 |
| chr11_+_10476851 | 0.69 |
ENST00000396553.2
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr5_+_149877440 | 0.68 |
ENST00000518299.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr4_+_159727272 | 0.68 |
ENST00000379346.3
|
FNIP2
|
folliculin interacting protein 2 |
| chr5_-_41794663 | 0.67 |
ENST00000510634.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr2_-_220117867 | 0.66 |
ENST00000456818.1
ENST00000447205.1 |
TUBA4A
|
tubulin, alpha 4a |
| chr10_+_127661942 | 0.65 |
ENST00000417114.1
ENST00000445510.1 ENST00000368691.1 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr11_-_128894053 | 0.65 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr11_-_8739383 | 0.63 |
ENST00000531060.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr16_+_53133070 | 0.63 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr19_-_36019123 | 0.62 |
ENST00000588674.1
ENST00000452271.2 ENST00000518157.1 |
SBSN
|
suprabasin |
| chr11_-_46848393 | 0.61 |
ENST00000526496.1
|
CKAP5
|
cytoskeleton associated protein 5 |
| chr11_-_66104237 | 0.61 |
ENST00000530056.1
|
RIN1
|
Ras and Rab interactor 1 |
| chr12_-_48152611 | 0.61 |
ENST00000389212.3
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr16_+_89988259 | 0.61 |
ENST00000554444.1
ENST00000556565.1 |
TUBB3
|
Tubulin beta-3 chain |
| chr1_-_6662919 | 0.60 |
ENST00000377658.4
ENST00000377663.3 |
KLHL21
|
kelch-like family member 21 |
| chr9_-_112179990 | 0.60 |
ENST00000394827.3
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr11_-_66103867 | 0.60 |
ENST00000424433.2
|
RIN1
|
Ras and Rab interactor 1 |
| chr4_+_170581213 | 0.60 |
ENST00000507875.1
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr12_-_118490403 | 0.60 |
ENST00000535496.1
|
WSB2
|
WD repeat and SOCS box containing 2 |
| chr16_+_57662596 | 0.59 |
ENST00000567397.1
ENST00000568979.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr1_+_223900034 | 0.59 |
ENST00000295006.5
|
CAPN2
|
calpain 2, (m/II) large subunit |
| chr11_+_59522837 | 0.59 |
ENST00000437946.2
|
STX3
|
syntaxin 3 |
| chr1_-_179112189 | 0.58 |
ENST00000512653.1
ENST00000344730.3 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr17_+_79651007 | 0.58 |
ENST00000572392.1
ENST00000577012.1 |
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr9_-_130637244 | 0.57 |
ENST00000373156.1
|
AK1
|
adenylate kinase 1 |
| chr11_-_66103932 | 0.57 |
ENST00000311320.4
|
RIN1
|
Ras and Rab interactor 1 |
| chr19_+_6740888 | 0.57 |
ENST00000596673.1
|
TRIP10
|
thyroid hormone receptor interactor 10 |
| chrX_-_129244336 | 0.56 |
ENST00000434609.1
|
ELF4
|
E74-like factor 4 (ets domain transcription factor) |
| chr2_+_220143989 | 0.56 |
ENST00000336576.5
|
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr8_-_29939933 | 0.56 |
ENST00000522794.1
|
TMEM66
|
transmembrane protein 66 |
| chr6_+_150070857 | 0.55 |
ENST00000544496.1
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr1_-_151965048 | 0.55 |
ENST00000368809.1
|
S100A10
|
S100 calcium binding protein A10 |
| chr1_-_157108130 | 0.55 |
ENST00000368192.4
|
ETV3
|
ets variant 3 |
| chr16_+_57662419 | 0.52 |
ENST00000388812.4
ENST00000538815.1 ENST00000456916.1 ENST00000567154.1 ENST00000388813.5 ENST00000562558.1 ENST00000566271.2 |
GPR56
|
G protein-coupled receptor 56 |
| chr10_+_121410882 | 0.52 |
ENST00000369085.3
|
BAG3
|
BCL2-associated athanogene 3 |
| chr17_+_65375082 | 0.52 |
ENST00000584471.1
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr17_+_79071365 | 0.52 |
ENST00000576756.1
|
BAIAP2
|
BAI1-associated protein 2 |
| chr19_+_49467232 | 0.52 |
ENST00000599784.1
ENST00000594305.1 |
CTD-2639E6.9
|
CTD-2639E6.9 |
| chr5_-_41794313 | 0.51 |
ENST00000512084.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr3_+_11178779 | 0.50 |
ENST00000438284.2
|
HRH1
|
histamine receptor H1 |
| chr7_+_143078379 | 0.49 |
ENST00000449630.1
ENST00000457235.1 |
ZYX
|
zyxin |
| chr5_+_150020214 | 0.49 |
ENST00000307662.4
|
SYNPO
|
synaptopodin |
| chr3_+_187930429 | 0.48 |
ENST00000420410.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr3_-_187454281 | 0.48 |
ENST00000232014.4
|
BCL6
|
B-cell CLL/lymphoma 6 |
| chr2_+_233562015 | 0.47 |
ENST00000427233.1
ENST00000373566.3 ENST00000373563.4 ENST00000428883.1 ENST00000456491.1 ENST00000409480.1 ENST00000421433.1 ENST00000425040.1 ENST00000430720.1 ENST00000409547.1 ENST00000423659.1 ENST00000409196.3 ENST00000409451.3 ENST00000429187.1 ENST00000440945.1 |
GIGYF2
|
GRB10 interacting GYF protein 2 |
| chr2_-_85641162 | 0.47 |
ENST00000447219.2
ENST00000409670.1 ENST00000409724.1 |
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr15_+_22892663 | 0.44 |
ENST00000313077.7
ENST00000561274.1 ENST00000560848.1 |
CYFIP1
|
cytoplasmic FMR1 interacting protein 1 |
| chr1_+_36554470 | 0.44 |
ENST00000373178.4
|
ADPRHL2
|
ADP-ribosylhydrolase like 2 |
| chr4_+_108746282 | 0.44 |
ENST00000503862.1
|
SGMS2
|
sphingomyelin synthase 2 |
| chr4_+_108745711 | 0.44 |
ENST00000394684.4
|
SGMS2
|
sphingomyelin synthase 2 |
| chr3_+_19988885 | 0.44 |
ENST00000422242.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr17_+_35851570 | 0.44 |
ENST00000394386.1
|
DUSP14
|
dual specificity phosphatase 14 |
| chr4_-_100356291 | 0.44 |
ENST00000476959.1
ENST00000482593.1 |
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr15_+_57211318 | 0.43 |
ENST00000557947.1
|
TCF12
|
transcription factor 12 |
| chr18_-_74207146 | 0.43 |
ENST00000443185.2
|
ZNF516
|
zinc finger protein 516 |
| chr4_-_146019693 | 0.42 |
ENST00000514390.1
|
ANAPC10
|
anaphase promoting complex subunit 10 |
| chrX_-_16730984 | 0.42 |
ENST00000380241.3
|
CTPS2
|
CTP synthase 2 |
| chr9_-_35112376 | 0.42 |
ENST00000488109.2
|
FAM214B
|
family with sequence similarity 214, member B |
| chr5_+_14664762 | 0.41 |
ENST00000284274.4
|
FAM105B
|
family with sequence similarity 105, member B |
| chr1_+_36621697 | 0.39 |
ENST00000373150.4
ENST00000373151.2 |
MAP7D1
|
MAP7 domain containing 1 |
| chr19_+_7953417 | 0.39 |
ENST00000600345.1
ENST00000598224.1 |
LRRC8E
|
leucine rich repeat containing 8 family, member E |
| chr17_+_28256874 | 0.38 |
ENST00000541045.1
ENST00000536908.2 |
EFCAB5
|
EF-hand calcium binding domain 5 |
| chr17_+_75315654 | 0.38 |
ENST00000590595.1
|
SEPT9
|
septin 9 |
| chr19_+_36630855 | 0.37 |
ENST00000589146.1
|
CAPNS1
|
calpain, small subunit 1 |
| chr2_-_96926313 | 0.36 |
ENST00000435268.1
|
TMEM127
|
transmembrane protein 127 |
| chr17_+_75315534 | 0.35 |
ENST00000590294.1
ENST00000329047.8 |
SEPT9
|
septin 9 |
| chr1_-_204121102 | 0.35 |
ENST00000367202.4
|
ETNK2
|
ethanolamine kinase 2 |
| chr9_+_34992846 | 0.34 |
ENST00000443266.1
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr3_-_98241713 | 0.33 |
ENST00000502288.1
ENST00000512147.1 ENST00000510541.1 ENST00000503621.1 ENST00000511081.1 |
CLDND1
|
claudin domain containing 1 |
| chr1_-_204121013 | 0.33 |
ENST00000367201.3
|
ETNK2
|
ethanolamine kinase 2 |
| chr7_+_116312411 | 0.33 |
ENST00000456159.1
ENST00000397752.3 ENST00000318493.6 |
MET
|
met proto-oncogene |
| chr7_-_35734730 | 0.33 |
ENST00000396081.1
ENST00000311350.3 |
HERPUD2
|
HERPUD family member 2 |
| chr15_+_44829334 | 0.32 |
ENST00000535391.1
|
EIF3J
|
eukaryotic translation initiation factor 3, subunit J |
| chr12_-_105352047 | 0.31 |
ENST00000432951.1
ENST00000415674.1 ENST00000424946.1 |
SLC41A2
|
solute carrier family 41 (magnesium transporter), member 2 |
| chr7_-_35734607 | 0.31 |
ENST00000427455.1
|
HERPUD2
|
HERPUD family member 2 |
| chr6_+_150070831 | 0.30 |
ENST00000367380.5
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr12_-_105352080 | 0.30 |
ENST00000433540.1
|
SLC41A2
|
solute carrier family 41 (magnesium transporter), member 2 |
| chr17_-_39538550 | 0.30 |
ENST00000394001.1
|
KRT34
|
keratin 34 |
| chr6_+_44214824 | 0.30 |
ENST00000371646.5
ENST00000353801.3 |
HSP90AB1
|
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chr16_+_22308717 | 0.30 |
ENST00000299853.5
ENST00000564209.1 ENST00000565358.1 ENST00000418581.2 ENST00000564883.1 ENST00000359210.4 ENST00000563024.1 |
POLR3E
|
polymerase (RNA) III (DNA directed) polypeptide E (80kD) |
| chr9_-_114246635 | 0.30 |
ENST00000338205.5
|
KIAA0368
|
KIAA0368 |
| chrX_-_16730688 | 0.30 |
ENST00000359276.4
|
CTPS2
|
CTP synthase 2 |
| chr19_-_51538148 | 0.29 |
ENST00000319590.4
ENST00000250351.4 |
KLK12
|
kallikrein-related peptidase 12 |
| chrX_+_3189861 | 0.29 |
ENST00000457435.1
ENST00000420429.2 |
CXorf28
|
chromosome X open reading frame 28 |
| chr12_-_118490217 | 0.28 |
ENST00000542304.1
|
WSB2
|
WD repeat and SOCS box containing 2 |
| chr7_-_150721570 | 0.28 |
ENST00000377974.2
ENST00000444312.1 ENST00000605938.1 ENST00000605952.1 |
ATG9B
|
autophagy related 9B |
| chr2_+_201980827 | 0.28 |
ENST00000309955.3
ENST00000443227.1 ENST00000341222.6 ENST00000355558.4 ENST00000340870.5 ENST00000341582.6 |
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr22_+_22764088 | 0.27 |
ENST00000390299.2
|
IGLV1-40
|
immunoglobulin lambda variable 1-40 |
| chr3_+_187930491 | 0.27 |
ENST00000443217.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr3_+_69134080 | 0.27 |
ENST00000273258.3
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr1_-_157108266 | 0.27 |
ENST00000326786.4
|
ETV3
|
ets variant 3 |
| chr19_+_50706866 | 0.27 |
ENST00000440075.2
ENST00000376970.2 ENST00000425460.1 ENST00000599920.1 ENST00000601313.1 |
MYH14
|
myosin, heavy chain 14, non-muscle |
| chr19_-_51538118 | 0.27 |
ENST00000529888.1
|
KLK12
|
kallikrein-related peptidase 12 |
| chr22_+_20861858 | 0.26 |
ENST00000414658.1
ENST00000432052.1 ENST00000425759.2 ENST00000292733.7 ENST00000542773.1 ENST00000263205.7 ENST00000406969.1 ENST00000382974.2 |
MED15
|
mediator complex subunit 15 |
| chr17_-_18161870 | 0.25 |
ENST00000579294.1
ENST00000545457.2 ENST00000379450.4 ENST00000578558.1 |
FLII
|
flightless I homolog (Drosophila) |
| chr9_+_4985016 | 0.25 |
ENST00000539801.1
|
JAK2
|
Janus kinase 2 |
| chr1_-_204121298 | 0.25 |
ENST00000367199.2
|
ETNK2
|
ethanolamine kinase 2 |
| chr10_-_73611046 | 0.24 |
ENST00000394934.1
ENST00000394936.3 |
PSAP
|
prosaposin |
| chr14_-_51863853 | 0.24 |
ENST00000556762.1
|
RP11-255G12.3
|
RP11-255G12.3 |
| chr11_-_65667997 | 0.24 |
ENST00000312562.2
ENST00000534222.1 |
FOSL1
|
FOS-like antigen 1 |
| chr6_-_119031228 | 0.23 |
ENST00000392500.3
ENST00000368488.5 ENST00000434604.1 |
CEP85L
|
centrosomal protein 85kDa-like |
| chr10_+_85899196 | 0.22 |
ENST00000372134.3
|
GHITM
|
growth hormone inducible transmembrane protein |
| chr22_+_20862321 | 0.22 |
ENST00000541476.1
ENST00000438962.1 |
MED15
|
mediator complex subunit 15 |
| chr19_+_7953384 | 0.22 |
ENST00000306708.6
|
LRRC8E
|
leucine rich repeat containing 8 family, member E |
| chr17_-_27467418 | 0.22 |
ENST00000528564.1
|
MYO18A
|
myosin XVIIIA |
| chr2_-_65593784 | 0.21 |
ENST00000443619.2
|
SPRED2
|
sprouty-related, EVH1 domain containing 2 |
| chr18_+_32290218 | 0.21 |
ENST00000348997.5
ENST00000588949.1 ENST00000597599.1 |
DTNA
|
dystrobrevin, alpha |
Network of associatons between targets according to the STRING database.
First level regulatory network of BACH2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.5 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.5 | 4.7 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.5 | 2.5 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.5 | 2.0 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.4 | 8.5 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.4 | 1.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.3 | 2.2 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.3 | 1.5 | GO:0030807 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.3 | 1.7 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.3 | 1.9 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.2 | 1.6 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.2 | 1.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.2 | 1.8 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.2 | 4.3 | GO:0016102 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.2 | 2.6 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.2 | 0.7 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.2 | 6.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 0.8 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 | 1.0 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.2 | 3.8 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.2 | 0.9 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 | 3.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 2.2 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.1 | 1.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.7 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 1.0 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 3.6 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 0.5 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.1 | 3.0 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 1.6 | GO:0007320 | insemination(GO:0007320) |
| 0.1 | 1.5 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.8 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 0.6 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.8 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.1 | 0.4 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.1 | 0.3 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.1 | 0.4 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.1 | 0.9 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.6 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 0.4 | GO:1902728 | positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.1 | 3.8 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.1 | 0.9 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 1.0 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 0.5 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.1 | 1.2 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.1 | 0.4 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 3.8 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.4 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.8 | GO:0097012 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.1 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.0 | 0.3 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.3 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.4 | GO:0032960 | regulation of inositol trisphosphate biosynthetic process(GO:0032960) positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.8 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.9 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.2 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 1.5 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.0 | 0.3 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.6 | GO:0098902 | regulation of membrane depolarization during action potential(GO:0098902) |
| 0.0 | 0.2 | GO:0060742 | epithelial cell differentiation involved in prostate gland development(GO:0060742) |
| 0.0 | 0.9 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.1 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 1.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.1 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 2.6 | GO:0030834 | regulation of actin filament depolymerization(GO:0030834) |
| 0.0 | 0.6 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.2 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.0 | 0.2 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.3 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.7 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.0 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.0 | 2.5 | GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) |
| 0.0 | 0.7 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.8 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 1.6 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.4 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.0 | 0.5 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.0 | 0.1 | GO:0010979 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) vitamin D catabolic process(GO:0042369) positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 1.1 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.2 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.2 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.8 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 1.6 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.6 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.5 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.7 | GO:0006936 | muscle contraction(GO:0006936) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.6 | 6.3 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.6 | 1.7 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.2 | 0.9 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.2 | 1.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.2 | 3.6 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.2 | 1.0 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.6 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.1 | 0.8 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 0.4 | GO:0043257 | laminin-8 complex(GO:0043257) |
| 0.1 | 7.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 3.0 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 0.9 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 1.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.6 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 1.6 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.1 | 0.5 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 2.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.8 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.7 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.8 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.3 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 1.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.4 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 1.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.8 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 2.0 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 2.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.5 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 1.0 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.0 | 0.4 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 2.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.7 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 2.0 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 2.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 2.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.5 | 3.6 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.4 | 2.6 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.4 | 1.1 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.3 | 1.2 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.2 | 0.8 | GO:1990175 | EH domain binding(GO:1990175) |
| 0.2 | 1.4 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 0.7 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.2 | 1.6 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 1.7 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 0.7 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 2.6 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 0.9 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.9 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 3.0 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 5.1 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 2.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.6 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 2.0 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 1.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.9 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.1 | 1.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.3 | GO:0002134 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.1 | 0.8 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.1 | 0.6 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 2.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 2.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 2.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 12.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.6 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.5 | GO:0008526 | phosphatidylcholine transporter activity(GO:0008525) phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.8 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 3.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.8 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.6 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 1.5 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 9.7 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 5.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 3.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 1.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.2 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 8.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 1.4 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 5.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 4.0 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.7 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 1.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.7 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 2.8 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.9 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.4 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 3.9 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 7.2 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 2.9 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.9 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.4 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |