Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
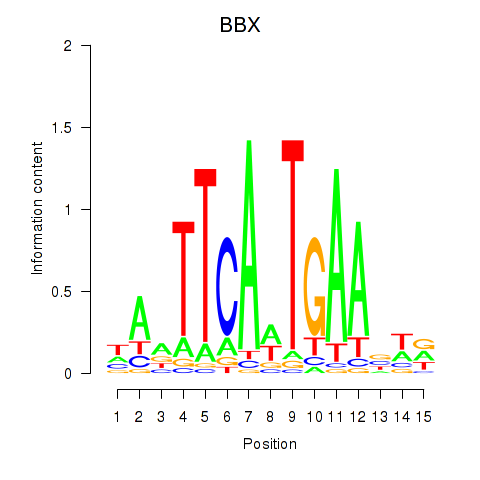
Results for BBX
Z-value: 0.71
Transcription factors associated with BBX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BBX
|
ENSG00000114439.14 | BBX high mobility group box domain containing |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BBX | hg19_v2_chr3_+_107241882_107241904 | -0.88 | 3.9e-07 | Click! |
Activity profile of BBX motif
Sorted Z-values of BBX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_155484155 | 4.86 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr4_+_155484103 | 4.64 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr6_-_25874440 | 1.35 |
ENST00000361703.6
ENST00000397060.4 |
SLC17A3
|
solute carrier family 17 (organic anion transporter), member 3 |
| chr9_-_116837249 | 0.93 |
ENST00000466610.2
|
AMBP
|
alpha-1-microglobulin/bikunin precursor |
| chr19_+_6887571 | 0.84 |
ENST00000250572.8
ENST00000381407.5 ENST00000312053.4 ENST00000450315.3 ENST00000381404.4 |
EMR1
|
egf-like module containing, mucin-like, hormone receptor-like 1 |
| chr19_+_9361606 | 0.77 |
ENST00000456448.1
|
OR7E24
|
olfactory receptor, family 7, subfamily E, member 24 |
| chr16_+_28763108 | 0.73 |
ENST00000357796.3
ENST00000550983.1 |
NPIPB9
|
nuclear pore complex interacting protein family, member B9 |
| chr6_-_107235331 | 0.68 |
ENST00000433965.1
ENST00000430094.1 |
RP1-60O19.1
|
RP1-60O19.1 |
| chr16_-_28374829 | 0.65 |
ENST00000532254.1
|
NPIPB6
|
nuclear pore complex interacting protein family, member B6 |
| chr6_-_107235287 | 0.64 |
ENST00000436659.1
ENST00000428750.1 ENST00000427903.1 |
RP1-60O19.1
|
RP1-60O19.1 |
| chr4_+_129349188 | 0.64 |
ENST00000511497.1
|
RP11-420A23.1
|
RP11-420A23.1 |
| chr5_+_140480083 | 0.62 |
ENST00000231130.2
|
PCDHB3
|
protocadherin beta 3 |
| chr10_-_94003003 | 0.60 |
ENST00000412050.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr5_+_140514782 | 0.59 |
ENST00000231134.5
|
PCDHB5
|
protocadherin beta 5 |
| chrX_+_54834004 | 0.57 |
ENST00000375068.1
|
MAGED2
|
melanoma antigen family D, 2 |
| chr7_-_16872932 | 0.57 |
ENST00000419572.2
ENST00000412973.1 |
AGR2
|
anterior gradient 2 |
| chr19_+_49109990 | 0.55 |
ENST00000321762.1
|
SPACA4
|
sperm acrosome associated 4 |
| chr5_+_74011328 | 0.54 |
ENST00000513336.1
|
HEXB
|
hexosaminidase B (beta polypeptide) |
| chr17_+_4853442 | 0.53 |
ENST00000522301.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr4_-_987217 | 0.50 |
ENST00000361661.2
ENST00000398516.2 |
SLC26A1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr12_-_10601963 | 0.49 |
ENST00000543893.1
|
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr4_+_88571429 | 0.48 |
ENST00000339673.6
ENST00000282479.7 |
DMP1
|
dentin matrix acidic phosphoprotein 1 |
| chr17_-_8263538 | 0.45 |
ENST00000535173.1
|
AC135178.1
|
HCG1985372; Uncharacterized protein; cDNA FLJ37541 fis, clone BRCAN2026340 |
| chr7_-_16921601 | 0.44 |
ENST00000402239.3
ENST00000310398.2 ENST00000414935.1 |
AGR3
|
anterior gradient 3 |
| chr14_+_23235886 | 0.44 |
ENST00000604262.1
ENST00000431881.2 ENST00000412791.1 ENST00000358043.5 |
OXA1L
|
oxidase (cytochrome c) assembly 1-like |
| chr20_-_30310656 | 0.44 |
ENST00000376055.4
|
BCL2L1
|
BCL2-like 1 |
| chr7_-_121944491 | 0.42 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr22_-_39928823 | 0.41 |
ENST00000334678.3
|
RPS19BP1
|
ribosomal protein S19 binding protein 1 |
| chr20_-_7921090 | 0.40 |
ENST00000378789.3
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr1_+_95616933 | 0.40 |
ENST00000604203.1
|
RP11-57H12.6
|
TMEM56-RWDD3 readthrough |
| chr17_-_16256718 | 0.40 |
ENST00000476243.1
ENST00000299736.4 |
CENPV
|
centromere protein V |
| chr20_-_30310693 | 0.39 |
ENST00000307677.4
ENST00000420653.1 |
BCL2L1
|
BCL2-like 1 |
| chr6_-_107235378 | 0.38 |
ENST00000606430.1
|
RP1-60O19.1
|
RP1-60O19.1 |
| chr14_-_23292596 | 0.37 |
ENST00000554741.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chrX_+_54834159 | 0.37 |
ENST00000375053.2
ENST00000347546.4 ENST00000375062.4 |
MAGED2
|
melanoma antigen family D, 2 |
| chr4_-_123542224 | 0.36 |
ENST00000264497.3
|
IL21
|
interleukin 21 |
| chr17_+_66255310 | 0.36 |
ENST00000448504.2
|
ARSG
|
arylsulfatase G |
| chr12_-_21487829 | 0.35 |
ENST00000445053.1
ENST00000452078.1 ENST00000458504.1 ENST00000422327.1 ENST00000421294.1 |
SLCO1A2
|
solute carrier organic anion transporter family, member 1A2 |
| chr3_+_111260856 | 0.35 |
ENST00000352690.4
|
CD96
|
CD96 molecule |
| chr1_+_170115142 | 0.35 |
ENST00000439373.2
|
METTL11B
|
methyltransferase like 11B |
| chr14_+_38264418 | 0.35 |
ENST00000267368.7
ENST00000382320.3 |
TTC6
|
tetratricopeptide repeat domain 6 |
| chr6_+_31895254 | 0.34 |
ENST00000299367.5
ENST00000442278.2 |
C2
|
complement component 2 |
| chr12_-_66035968 | 0.34 |
ENST00000537250.1
|
RP11-230G5.2
|
RP11-230G5.2 |
| chr16_+_56672571 | 0.30 |
ENST00000290705.8
|
MT1A
|
metallothionein 1A |
| chr2_+_197577841 | 0.30 |
ENST00000409270.1
|
CCDC150
|
coiled-coil domain containing 150 |
| chr2_-_158182322 | 0.30 |
ENST00000420719.2
ENST00000409216.1 |
ERMN
|
ermin, ERM-like protein |
| chr3_+_111260954 | 0.30 |
ENST00000283285.5
|
CD96
|
CD96 molecule |
| chr14_+_67656110 | 0.30 |
ENST00000524532.1
ENST00000530728.1 |
FAM71D
|
family with sequence similarity 71, member D |
| chr4_-_104021009 | 0.29 |
ENST00000509245.1
ENST00000296424.4 |
BDH2
|
3-hydroxybutyrate dehydrogenase, type 2 |
| chr7_+_117120106 | 0.28 |
ENST00000426809.1
|
CFTR
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr11_-_58035732 | 0.28 |
ENST00000395079.2
|
OR10W1
|
olfactory receptor, family 10, subfamily W, member 1 |
| chr1_+_229440129 | 0.27 |
ENST00000366688.3
|
SPHAR
|
S-phase response (cyclin related) |
| chr17_-_17184605 | 0.26 |
ENST00000268717.5
|
COPS3
|
COP9 signalosome subunit 3 |
| chr20_-_30310336 | 0.26 |
ENST00000434194.1
ENST00000376062.2 |
BCL2L1
|
BCL2-like 1 |
| chr17_-_59668550 | 0.26 |
ENST00000521764.1
|
NACA2
|
nascent polypeptide-associated complex alpha subunit 2 |
| chr9_-_93405352 | 0.26 |
ENST00000375765.3
|
DIRAS2
|
DIRAS family, GTP-binding RAS-like 2 |
| chr4_+_187813673 | 0.25 |
ENST00000606881.1
|
RP11-11N5.3
|
RP11-11N5.3 |
| chr18_-_56985776 | 0.25 |
ENST00000587244.1
|
CPLX4
|
complexin 4 |
| chr11_-_14541872 | 0.24 |
ENST00000419365.2
ENST00000530457.1 ENST00000532256.1 ENST00000533068.1 |
PSMA1
|
proteasome (prosome, macropain) subunit, alpha type, 1 |
| chr2_-_158182410 | 0.23 |
ENST00000419116.2
ENST00000410096.1 |
ERMN
|
ermin, ERM-like protein |
| chr17_+_40950900 | 0.23 |
ENST00000588527.1
|
CNTD1
|
cyclin N-terminal domain containing 1 |
| chr7_+_95171457 | 0.23 |
ENST00000601424.1
|
AC002451.1
|
Protein LOC100996577 |
| chr4_-_104020968 | 0.23 |
ENST00000504285.1
|
BDH2
|
3-hydroxybutyrate dehydrogenase, type 2 |
| chr3_-_112218378 | 0.23 |
ENST00000334529.5
|
BTLA
|
B and T lymphocyte associated |
| chr9_-_95244781 | 0.23 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr6_-_49712123 | 0.22 |
ENST00000263045.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr3_+_130159354 | 0.22 |
ENST00000373157.4
|
COL6A5
|
collagen, type VI, alpha 5 |
| chr9_-_95166884 | 0.22 |
ENST00000375561.5
|
OGN
|
osteoglycin |
| chr9_-_95166841 | 0.21 |
ENST00000262551.4
|
OGN
|
osteoglycin |
| chr2_+_110656005 | 0.21 |
ENST00000437679.2
|
LIMS3
|
LIM and senescent cell antigen-like domains 3 |
| chr1_-_216978709 | 0.21 |
ENST00000360012.3
|
ESRRG
|
estrogen-related receptor gamma |
| chr7_-_130080977 | 0.21 |
ENST00000223208.5
|
CEP41
|
centrosomal protein 41kDa |
| chr13_+_76413852 | 0.21 |
ENST00000533809.2
|
LMO7
|
LIM domain 7 |
| chr3_+_140981456 | 0.21 |
ENST00000504264.1
|
ACPL2
|
acid phosphatase-like 2 |
| chrX_-_119077695 | 0.20 |
ENST00000371410.3
|
NKAP
|
NFKB activating protein |
| chrX_-_52827141 | 0.20 |
ENST00000375511.3
|
SPANXN5
|
SPANX family, member N5 |
| chr3_+_177534653 | 0.20 |
ENST00000436078.1
|
RP11-91K9.1
|
RP11-91K9.1 |
| chr1_+_28527070 | 0.19 |
ENST00000596102.1
|
AL353354.2
|
AL353354.2 |
| chr10_+_90562487 | 0.19 |
ENST00000404743.4
|
LIPM
|
lipase, family member M |
| chr4_-_186696515 | 0.19 |
ENST00000456596.1
ENST00000414724.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr18_-_13915530 | 0.18 |
ENST00000327606.3
|
MC2R
|
melanocortin 2 receptor (adrenocorticotropic hormone) |
| chr15_+_54901540 | 0.18 |
ENST00000539562.2
|
UNC13C
|
unc-13 homolog C (C. elegans) |
| chr2_+_63816295 | 0.18 |
ENST00000539945.1
ENST00000544381.1 |
MDH1
|
malate dehydrogenase 1, NAD (soluble) |
| chr11_+_61717279 | 0.18 |
ENST00000378043.4
|
BEST1
|
bestrophin 1 |
| chr15_+_54305101 | 0.18 |
ENST00000260323.11
ENST00000545554.1 ENST00000537900.1 |
UNC13C
|
unc-13 homolog C (C. elegans) |
| chrX_-_140786896 | 0.17 |
ENST00000370515.3
|
SPANXD
|
SPANX family, member D |
| chr3_+_111260980 | 0.17 |
ENST00000438817.2
|
CD96
|
CD96 molecule |
| chr6_-_135818368 | 0.17 |
ENST00000367798.2
|
AHI1
|
Abelson helper integration site 1 |
| chr3_+_81043000 | 0.17 |
ENST00000482617.1
|
RP11-6B4.1
|
RP11-6B4.1 |
| chr12_+_96196875 | 0.16 |
ENST00000553095.1
|
RP11-536G4.1
|
Uncharacterized protein |
| chr10_-_4285923 | 0.16 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr12_-_8765446 | 0.16 |
ENST00000537228.1
ENST00000229335.6 |
AICDA
|
activation-induced cytidine deaminase |
| chrX_+_140677562 | 0.15 |
ENST00000370518.3
|
SPANXA2
|
SPANX family, member A2 |
| chr4_+_174818390 | 0.15 |
ENST00000509968.1
ENST00000512263.1 |
RP11-161D15.1
|
RP11-161D15.1 |
| chr18_+_63273310 | 0.15 |
ENST00000579862.1
|
RP11-775G23.1
|
RP11-775G23.1 |
| chr4_-_987164 | 0.15 |
ENST00000398520.2
|
SLC26A1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr10_-_4285835 | 0.14 |
ENST00000454470.1
|
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr6_+_72922505 | 0.14 |
ENST00000401910.3
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr7_+_117120017 | 0.14 |
ENST00000003084.6
ENST00000454343.1 |
CFTR
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr4_-_186696636 | 0.14 |
ENST00000444771.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr6_+_72922590 | 0.14 |
ENST00000523963.1
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr3_-_112218205 | 0.14 |
ENST00000383680.4
|
BTLA
|
B and T lymphocyte associated |
| chr11_+_61717336 | 0.13 |
ENST00000378042.3
|
BEST1
|
bestrophin 1 |
| chr14_+_22458631 | 0.13 |
ENST00000390444.1
|
TRAV16
|
T cell receptor alpha variable 16 |
| chr9_-_107361788 | 0.13 |
ENST00000374779.2
|
OR13C5
|
olfactory receptor, family 13, subfamily C, member 5 |
| chr4_-_186696561 | 0.13 |
ENST00000445115.1
ENST00000451701.1 ENST00000457247.1 ENST00000435480.1 ENST00000425679.1 ENST00000457934.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr4_+_114066764 | 0.13 |
ENST00000511380.1
|
ANK2
|
ankyrin 2, neuronal |
| chr2_+_13677795 | 0.13 |
ENST00000434509.1
|
AC092635.1
|
AC092635.1 |
| chr2_-_120124258 | 0.13 |
ENST00000409877.1
ENST00000409523.1 ENST00000409466.2 ENST00000414534.1 |
C2orf76
|
chromosome 2 open reading frame 76 |
| chr8_-_28347737 | 0.13 |
ENST00000517673.1
ENST00000518734.1 ENST00000346498.2 ENST00000380254.2 |
FBXO16
|
F-box protein 16 |
| chr1_+_109265033 | 0.13 |
ENST00000445274.1
|
FNDC7
|
fibronectin type III domain containing 7 |
| chr8_-_116673894 | 0.12 |
ENST00000395713.2
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr6_-_133084580 | 0.11 |
ENST00000525270.1
ENST00000530536.1 ENST00000524919.1 |
VNN2
|
vanin 2 |
| chr6_-_133084564 | 0.11 |
ENST00000532012.1
|
VNN2
|
vanin 2 |
| chr4_-_186734275 | 0.11 |
ENST00000456060.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr2_+_228735763 | 0.10 |
ENST00000373666.2
|
DAW1
|
dynein assembly factor with WDR repeat domains 1 |
| chr6_-_88001706 | 0.10 |
ENST00000369576.2
|
GJB7
|
gap junction protein, beta 7, 25kDa |
| chr10_+_115674530 | 0.09 |
ENST00000451472.1
|
AL162407.1
|
CDNA FLJ20147 fis, clone COL07954; HCG1781466; Uncharacterized protein |
| chr12_-_11339543 | 0.09 |
ENST00000334266.1
|
TAS2R42
|
taste receptor, type 2, member 42 |
| chr7_+_128349106 | 0.09 |
ENST00000485070.1
|
FAM71F1
|
family with sequence similarity 71, member F1 |
| chr4_-_70080449 | 0.09 |
ENST00000446444.1
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr19_+_31658405 | 0.08 |
ENST00000589511.1
|
CTC-439O9.3
|
CTC-439O9.3 |
| chr3_+_51851612 | 0.08 |
ENST00000456080.1
|
IQCF3
|
IQ motif containing F3 |
| chr13_+_28494130 | 0.07 |
ENST00000381033.4
|
PDX1
|
pancreatic and duodenal homeobox 1 |
| chr13_+_50570019 | 0.07 |
ENST00000442421.1
|
TRIM13
|
tripartite motif containing 13 |
| chr12_-_66036137 | 0.07 |
ENST00000539653.1
ENST00000544557.1 |
RP11-230G5.2
|
RP11-230G5.2 |
| chr6_+_151358048 | 0.07 |
ENST00000450635.1
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chrX_+_144328348 | 0.06 |
ENST00000370493.3
|
SPANXN1
|
SPANX family, member N1 |
| chr6_+_31895287 | 0.06 |
ENST00000447952.2
|
C2
|
complement component 2 |
| chr4_-_186697044 | 0.05 |
ENST00000437304.2
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr6_+_53948328 | 0.05 |
ENST00000370876.2
|
MLIP
|
muscular LMNA-interacting protein |
| chr1_-_153123345 | 0.05 |
ENST00000368748.4
|
SPRR2G
|
small proline-rich protein 2G |
| chr4_-_186696425 | 0.05 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr6_+_29424958 | 0.05 |
ENST00000377136.1
ENST00000377133.1 |
OR2H1
|
olfactory receptor, family 2, subfamily H, member 1 |
| chr2_-_231084617 | 0.05 |
ENST00000409815.2
|
SP110
|
SP110 nuclear body protein |
| chr6_+_54173227 | 0.05 |
ENST00000259782.4
ENST00000370864.3 |
TINAG
|
tubulointerstitial nephritis antigen |
| chr12_-_100378006 | 0.05 |
ENST00000547776.2
ENST00000329257.7 ENST00000547010.1 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr12_-_100656134 | 0.05 |
ENST00000548313.1
|
DEPDC4
|
DEP domain containing 4 |
| chr5_+_180794269 | 0.04 |
ENST00000456475.1
|
OR4F3
|
olfactory receptor, family 4, subfamily F, member 3 |
| chr1_-_206785789 | 0.04 |
ENST00000437518.1
ENST00000367114.3 |
EIF2D
|
eukaryotic translation initiation factor 2D |
| chr7_+_135611542 | 0.04 |
ENST00000416501.1
|
AC015987.2
|
AC015987.2 |
| chr5_+_140810132 | 0.04 |
ENST00000252085.3
|
PCDHGA12
|
protocadherin gamma subfamily A, 12 |
| chr6_-_130543958 | 0.04 |
ENST00000437477.2
ENST00000439090.2 |
SAMD3
|
sterile alpha motif domain containing 3 |
| chr5_-_119669160 | 0.03 |
ENST00000514240.1
|
CTC-552D5.1
|
CTC-552D5.1 |
| chr10_-_85985294 | 0.03 |
ENST00000538192.1
|
LRIT2
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 2 |
| chr10_-_85985282 | 0.03 |
ENST00000372113.4
|
LRIT2
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 2 |
| chr11_+_22689648 | 0.03 |
ENST00000278187.3
|
GAS2
|
growth arrest-specific 2 |
| chr6_+_53948221 | 0.03 |
ENST00000460844.2
|
MLIP
|
muscular LMNA-interacting protein |
| chr6_-_161085291 | 0.03 |
ENST00000316300.5
|
LPA
|
lipoprotein, Lp(a) |
| chr2_+_12246664 | 0.03 |
ENST00000449986.1
|
AC096559.1
|
AC096559.1 |
| chr8_-_108510224 | 0.02 |
ENST00000517746.1
ENST00000297450.3 |
ANGPT1
|
angiopoietin 1 |
| chr12_-_99548270 | 0.02 |
ENST00000546568.1
ENST00000332712.7 ENST00000546960.1 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chrX_-_102531717 | 0.02 |
ENST00000372680.1
|
TCEAL5
|
transcription elongation factor A (SII)-like 5 |
| chr13_-_81801115 | 0.02 |
ENST00000567258.1
|
LINC00564
|
long intergenic non-protein coding RNA 564 |
| chr16_-_8622226 | 0.02 |
ENST00000568335.1
|
TMEM114
|
transmembrane protein 114 |
| chr20_+_23420885 | 0.01 |
ENST00000246020.2
|
CSTL1
|
cystatin-like 1 |
| chr12_+_50690489 | 0.01 |
ENST00000598429.1
|
AC140061.12
|
Uncharacterized protein |
| chr1_-_36948879 | 0.01 |
ENST00000373106.1
ENST00000373104.1 ENST00000373103.1 |
CSF3R
|
colony stimulating factor 3 receptor (granulocyte) |
| chr13_+_76445187 | 0.00 |
ENST00000318245.4
|
C13orf45
|
chromosome 13 open reading frame 45 |
| chr1_+_110577229 | 0.00 |
ENST00000369795.3
ENST00000369794.2 |
STRIP1
|
striatin interacting protein 1 |
| chr4_-_186682716 | 0.00 |
ENST00000445343.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr5_+_179921344 | 0.00 |
ENST00000261951.4
|
CNOT6
|
CCR4-NOT transcription complex, subunit 6 |
| chr16_-_31564881 | 0.00 |
ENST00000567765.1
|
CTD-2014E2.6
|
CTD-2014E2.6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of BBX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 9.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.3 | 0.8 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.2 | 0.6 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 | 1.1 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.1 | 1.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.4 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.1 | 0.4 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.1 | 0.5 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.1 | 0.4 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 0.3 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.1 | 0.6 | GO:1903899 | lung goblet cell differentiation(GO:0060480) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.1 | 0.9 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.5 | GO:0007619 | courtship behavior(GO:0007619) |
| 0.1 | 0.4 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.9 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.7 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 | 0.4 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.1 | 0.2 | GO:0070121 | pronephric nephron morphogenesis(GO:0039007) pronephric nephron tubule morphogenesis(GO:0039008) pronephric duct development(GO:0039022) pronephric duct morphogenesis(GO:0039023) Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.5 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.2 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.4 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 0.5 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.4 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.4 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.4 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.3 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.2 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.1 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.6 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 1.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 9.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 1.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.4 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.1 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.3 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 1.3 | GO:0031526 | brush border membrane(GO:0031526) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0019862 | IgA binding(GO:0019862) |
| 0.2 | 1.3 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.4 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.1 | 0.5 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.4 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.1 | 0.5 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 1.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.3 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.1 | 0.5 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 9.5 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 0.4 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.5 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.2 | GO:0047860 | diiodophenylpyruvate reductase activity(GO:0047860) |
| 0.0 | 0.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.7 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.9 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.6 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.4 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 9.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 9.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 1.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.1 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.3 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 1.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |