Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
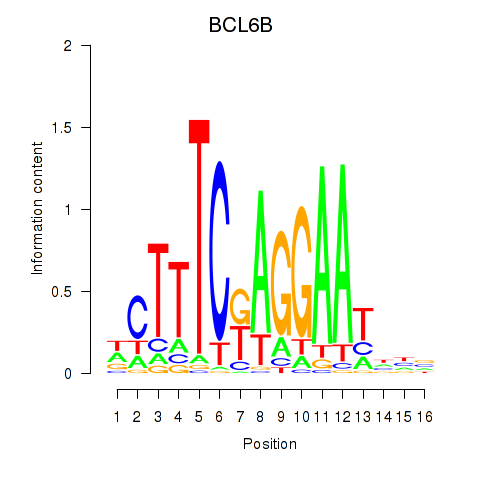
Results for BCL6B
Z-value: 0.37
Transcription factors associated with BCL6B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BCL6B
|
ENSG00000161940.6 | BCL6B transcription repressor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BCL6B | hg19_v2_chr17_+_6926339_6926377 | -0.16 | 5.0e-01 | Click! |
Activity profile of BCL6B motif
Sorted Z-values of BCL6B motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_10007448 | 0.94 |
ENST00000538152.1
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr14_+_95078714 | 0.76 |
ENST00000393078.3
ENST00000393080.4 ENST00000467132.1 |
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr12_+_8975061 | 0.65 |
ENST00000299698.7
|
A2ML1
|
alpha-2-macroglobulin-like 1 |
| chr11_+_123986069 | 0.61 |
ENST00000456829.2
ENST00000361352.5 ENST00000449321.1 ENST00000392748.1 ENST00000360334.4 ENST00000392744.4 |
VWA5A
|
von Willebrand factor A domain containing 5A |
| chr19_-_460996 | 0.56 |
ENST00000264554.6
|
SHC2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr13_-_33112823 | 0.51 |
ENST00000504114.1
|
N4BP2L2
|
NEDD4 binding protein 2-like 2 |
| chr1_+_209878182 | 0.49 |
ENST00000367027.3
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr12_-_123187890 | 0.47 |
ENST00000328880.5
|
HCAR2
|
hydroxycarboxylic acid receptor 2 |
| chr19_+_10197463 | 0.46 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr3_-_50649192 | 0.45 |
ENST00000443053.2
ENST00000348721.3 |
CISH
|
cytokine inducible SH2-containing protein |
| chr4_+_69313145 | 0.43 |
ENST00000305363.4
|
TMPRSS11E
|
transmembrane protease, serine 11E |
| chr13_-_33112899 | 0.41 |
ENST00000267068.3
ENST00000357505.6 ENST00000399396.3 |
N4BP2L2
|
NEDD4 binding protein 2-like 2 |
| chr12_-_123201337 | 0.40 |
ENST00000528880.2
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr11_-_18258342 | 0.40 |
ENST00000278222.4
|
SAA4
|
serum amyloid A4, constitutive |
| chr11_+_18230727 | 0.39 |
ENST00000527059.1
|
RP11-113D6.10
|
Putative mitochondrial carrier protein LOC494141 |
| chr3_+_177534653 | 0.36 |
ENST00000436078.1
|
RP11-91K9.1
|
RP11-91K9.1 |
| chr2_-_216257849 | 0.36 |
ENST00000456923.1
|
FN1
|
fibronectin 1 |
| chr12_-_15082050 | 0.34 |
ENST00000540097.1
|
ERP27
|
endoplasmic reticulum protein 27 |
| chr11_+_120081475 | 0.32 |
ENST00000328965.4
|
OAF
|
OAF homolog (Drosophila) |
| chr1_-_153321301 | 0.30 |
ENST00000368739.3
|
PGLYRP4
|
peptidoglycan recognition protein 4 |
| chr19_-_4831701 | 0.30 |
ENST00000248244.5
|
TICAM1
|
toll-like receptor adaptor molecule 1 |
| chr6_-_28304152 | 0.30 |
ENST00000435857.1
|
ZSCAN31
|
zinc finger and SCAN domain containing 31 |
| chr6_-_28303901 | 0.28 |
ENST00000439158.1
ENST00000446474.1 ENST00000414431.1 ENST00000344279.6 ENST00000453745.1 |
ZSCAN31
|
zinc finger and SCAN domain containing 31 |
| chr9_-_117880477 | 0.28 |
ENST00000534839.1
ENST00000340094.3 ENST00000535648.1 ENST00000346706.3 ENST00000345230.3 ENST00000350763.4 |
TNC
|
tenascin C |
| chr6_+_7541845 | 0.28 |
ENST00000418664.2
|
DSP
|
desmoplakin |
| chr6_-_28973037 | 0.27 |
ENST00000377179.3
|
ZNF311
|
zinc finger protein 311 |
| chr12_+_104324112 | 0.25 |
ENST00000299767.5
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr3_+_102153859 | 0.25 |
ENST00000306176.1
ENST00000466937.1 |
ZPLD1
|
zona pellucida-like domain containing 1 |
| chr10_-_70287231 | 0.24 |
ENST00000609923.1
|
SLC25A16
|
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
| chr14_+_21566980 | 0.24 |
ENST00000418511.2
ENST00000554329.2 |
TMEM253
|
transmembrane protein 253 |
| chr16_+_16481598 | 0.24 |
ENST00000327792.5
|
NPIPA7
|
nuclear pore complex interacting protein family, member A7 |
| chr1_-_230850043 | 0.23 |
ENST00000366667.4
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8) |
| chr2_+_120189422 | 0.23 |
ENST00000306406.4
|
TMEM37
|
transmembrane protein 37 |
| chr16_-_66952779 | 0.21 |
ENST00000570262.1
ENST00000394055.3 ENST00000299752.4 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr16_-_66952742 | 0.21 |
ENST00000565235.2
ENST00000568632.1 ENST00000565796.1 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr10_-_62493223 | 0.20 |
ENST00000373827.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr4_-_76957214 | 0.20 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chr6_+_7541808 | 0.20 |
ENST00000379802.3
|
DSP
|
desmoplakin |
| chr12_-_42877726 | 0.20 |
ENST00000548696.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr4_-_153303658 | 0.19 |
ENST00000296555.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr12_-_122238913 | 0.19 |
ENST00000537157.1
|
AC084018.1
|
AC084018.1 |
| chr19_+_39786962 | 0.19 |
ENST00000333625.2
|
IFNL1
|
interferon, lambda 1 |
| chr7_+_21582638 | 0.19 |
ENST00000409508.3
ENST00000328843.6 |
DNAH11
|
dynein, axonemal, heavy chain 11 |
| chr8_-_130799134 | 0.19 |
ENST00000276708.4
|
GSDMC
|
gasdermin C |
| chr1_-_151826173 | 0.18 |
ENST00000368817.5
|
THEM5
|
thioesterase superfamily member 5 |
| chr5_-_145214848 | 0.18 |
ENST00000505416.1
ENST00000334744.4 ENST00000358004.2 ENST00000511435.1 |
PRELID2
|
PRELI domain containing 2 |
| chr17_+_76183398 | 0.18 |
ENST00000409257.5
|
AFMID
|
arylformamidase |
| chr3_+_44771088 | 0.18 |
ENST00000396048.2
|
ZNF501
|
zinc finger protein 501 |
| chr6_+_42584847 | 0.17 |
ENST00000372883.3
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr5_+_140501581 | 0.17 |
ENST00000194152.1
|
PCDHB4
|
protocadherin beta 4 |
| chr13_+_20207782 | 0.17 |
ENST00000414242.2
ENST00000361479.5 |
MPHOSPH8
|
M-phase phosphoprotein 8 |
| chr12_-_42877764 | 0.17 |
ENST00000455697.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr7_+_74072011 | 0.16 |
ENST00000324896.4
ENST00000353920.4 ENST00000346152.4 ENST00000416070.1 |
GTF2I
|
general transcription factor IIi |
| chr3_-_187455680 | 0.16 |
ENST00000438077.1
|
BCL6
|
B-cell CLL/lymphoma 6 |
| chr6_-_83775442 | 0.16 |
ENST00000369745.5
|
UBE3D
|
ubiquitin protein ligase E3D |
| chr10_-_81708854 | 0.16 |
ENST00000372292.3
|
SFTPD
|
surfactant protein D |
| chr4_-_184241927 | 0.16 |
ENST00000323319.5
|
CLDN22
|
claudin 22 |
| chr15_+_45003675 | 0.15 |
ENST00000558401.1
ENST00000559916.1 ENST00000544417.1 |
B2M
|
beta-2-microglobulin |
| chr13_-_33112956 | 0.15 |
ENST00000505213.1
|
N4BP2L2
|
NEDD4 binding protein 2-like 2 |
| chr2_+_169659121 | 0.14 |
ENST00000397206.2
ENST00000397209.2 ENST00000421711.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr3_+_156799587 | 0.14 |
ENST00000469196.1
|
RP11-6F2.5
|
RP11-6F2.5 |
| chr13_+_53191605 | 0.14 |
ENST00000342657.3
ENST00000398039.1 |
HNRNPA1L2
|
heterogeneous nuclear ribonucleoprotein A1-like 2 |
| chr16_+_14396121 | 0.14 |
ENST00000570945.1
|
RP11-65J21.3
|
RP11-65J21.3 |
| chr10_-_70287172 | 0.14 |
ENST00000539557.1
|
SLC25A16
|
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
| chr5_-_172198190 | 0.13 |
ENST00000239223.3
|
DUSP1
|
dual specificity phosphatase 1 |
| chr12_-_11002063 | 0.13 |
ENST00000544994.1
ENST00000228811.4 ENST00000540107.1 |
PRR4
|
proline rich 4 (lacrimal) |
| chr14_+_75988851 | 0.13 |
ENST00000555504.1
|
BATF
|
basic leucine zipper transcription factor, ATF-like |
| chr10_+_11207438 | 0.13 |
ENST00000609692.1
ENST00000354897.3 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr6_+_31588478 | 0.13 |
ENST00000376007.4
ENST00000376033.2 |
PRRC2A
|
proline-rich coiled-coil 2A |
| chr7_+_23719749 | 0.13 |
ENST00000409192.3
ENST00000344962.4 ENST00000409653.1 ENST00000409994.3 |
FAM221A
|
family with sequence similarity 221, member A |
| chr12_-_48276710 | 0.12 |
ENST00000550314.1
|
VDR
|
vitamin D (1,25- dihydroxyvitamin D3) receptor |
| chr5_-_145214893 | 0.12 |
ENST00000394450.2
|
PRELID2
|
PRELI domain containing 2 |
| chr1_-_31845914 | 0.12 |
ENST00000373713.2
|
FABP3
|
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) |
| chr5_-_38845812 | 0.12 |
ENST00000513480.1
ENST00000512519.1 |
CTD-2127H9.1
|
CTD-2127H9.1 |
| chr4_-_155511887 | 0.12 |
ENST00000302053.3
ENST00000403106.3 |
FGA
|
fibrinogen alpha chain |
| chr19_-_13227463 | 0.12 |
ENST00000437766.1
ENST00000221504.8 |
TRMT1
|
tRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr3_-_23958402 | 0.11 |
ENST00000415901.2
ENST00000416026.2 ENST00000412028.1 ENST00000388759.3 ENST00000437230.1 |
NKIRAS1
|
NFKB inhibitor interacting Ras-like 1 |
| chr5_+_72251793 | 0.11 |
ENST00000430046.2
ENST00000341845.6 |
FCHO2
|
FCH domain only 2 |
| chr15_-_49913126 | 0.11 |
ENST00000561064.1
ENST00000299338.6 |
FAM227B
|
family with sequence similarity 227, member B |
| chr11_+_26495447 | 0.11 |
ENST00000531568.1
|
ANO3
|
anoctamin 3 |
| chr17_+_4981535 | 0.11 |
ENST00000318833.3
|
ZFP3
|
ZFP3 zinc finger protein |
| chr1_-_150669500 | 0.11 |
ENST00000271732.3
|
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr11_+_58912240 | 0.10 |
ENST00000527629.1
ENST00000361723.3 ENST00000531408.1 |
FAM111A
|
family with sequence similarity 111, member A |
| chr4_+_17616253 | 0.10 |
ENST00000237380.7
|
MED28
|
mediator complex subunit 28 |
| chr13_-_41345277 | 0.10 |
ENST00000323563.6
|
MRPS31
|
mitochondrial ribosomal protein S31 |
| chr20_+_58251716 | 0.10 |
ENST00000355648.4
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr3_-_23958506 | 0.10 |
ENST00000425478.2
|
NKIRAS1
|
NFKB inhibitor interacting Ras-like 1 |
| chr17_+_47439733 | 0.10 |
ENST00000507337.1
|
RP11-1079K10.3
|
RP11-1079K10.3 |
| chr12_+_19358228 | 0.10 |
ENST00000424268.1
ENST00000543806.1 |
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr11_+_107643129 | 0.10 |
ENST00000447610.1
|
AP001024.2
|
Uncharacterized protein |
| chr17_+_79761997 | 0.10 |
ENST00000400723.3
ENST00000570996.1 |
GCGR
|
glucagon receptor |
| chr22_-_50699701 | 0.10 |
ENST00000395780.1
|
MAPK12
|
mitogen-activated protein kinase 12 |
| chr11_+_64059464 | 0.10 |
ENST00000394525.2
|
KCNK4
|
potassium channel, subfamily K, member 4 |
| chr18_-_52989217 | 0.09 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr6_-_45544507 | 0.09 |
ENST00000563807.1
|
RP1-166H4.2
|
RP1-166H4.2 |
| chr2_+_70121075 | 0.09 |
ENST00000409116.1
|
SNRNP27
|
small nuclear ribonucleoprotein 27kDa (U4/U6.U5) |
| chr13_+_44596471 | 0.09 |
ENST00000592085.1
ENST00000591799.1 |
LINC00284
|
long intergenic non-protein coding RNA 284 |
| chrX_+_134847370 | 0.09 |
ENST00000482795.1
|
CT45A1
|
cancer/testis antigen family 45, member A1 |
| chr11_+_18230685 | 0.09 |
ENST00000340135.3
ENST00000534640.1 |
RP11-113D6.10
|
Putative mitochondrial carrier protein LOC494141 |
| chr13_-_28194541 | 0.09 |
ENST00000316334.3
|
LNX2
|
ligand of numb-protein X 2 |
| chr4_+_86396265 | 0.09 |
ENST00000395184.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr3_+_23958470 | 0.09 |
ENST00000434031.2
ENST00000413699.1 ENST00000456530.2 |
RPL15
|
ribosomal protein L15 |
| chr2_-_31030277 | 0.09 |
ENST00000534090.2
ENST00000295055.8 |
CAPN13
|
calpain 13 |
| chrX_-_134936550 | 0.08 |
ENST00000494421.1
|
CT45A4
|
cancer/testis antigen family 45, member A4 |
| chr17_+_66539369 | 0.08 |
ENST00000600820.1
|
AC079210.1
|
Uncharacterized protein; cDNA FLJ45097 fis, clone BRAWH3031054 |
| chr3_+_23959185 | 0.08 |
ENST00000354811.5
|
RPL15
|
ribosomal protein L15 |
| chr17_-_54893250 | 0.08 |
ENST00000397862.2
|
C17orf67
|
chromosome 17 open reading frame 67 |
| chr17_+_76183432 | 0.08 |
ENST00000591256.1
ENST00000589256.1 ENST00000588800.1 ENST00000591952.1 ENST00000327898.5 ENST00000586542.1 ENST00000586731.1 |
AFMID
|
arylformamidase |
| chr3_+_185046676 | 0.08 |
ENST00000428617.1
ENST00000443863.1 |
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr6_+_125540951 | 0.08 |
ENST00000524679.1
|
TPD52L1
|
tumor protein D52-like 1 |
| chr4_+_86699834 | 0.08 |
ENST00000395183.2
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr12_+_70132458 | 0.08 |
ENST00000247833.7
|
RAB3IP
|
RAB3A interacting protein |
| chr12_+_19358192 | 0.07 |
ENST00000538305.1
|
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr7_+_28452130 | 0.07 |
ENST00000357727.2
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr18_-_52989525 | 0.07 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr20_+_56136136 | 0.07 |
ENST00000319441.4
ENST00000543666.1 |
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr22_-_31503490 | 0.07 |
ENST00000400299.2
|
SELM
|
Selenoprotein M |
| chr12_+_31812121 | 0.07 |
ENST00000395763.3
|
METTL20
|
methyltransferase like 20 |
| chr3_+_35683651 | 0.07 |
ENST00000187397.4
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr3_+_46618727 | 0.07 |
ENST00000296145.5
|
TDGF1
|
teratocarcinoma-derived growth factor 1 |
| chr12_+_93096619 | 0.07 |
ENST00000397833.3
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chrX_+_135570046 | 0.07 |
ENST00000370648.3
|
BRS3
|
bombesin-like receptor 3 |
| chr20_-_18447667 | 0.07 |
ENST00000262547.5
ENST00000329494.5 ENST00000357236.4 |
DZANK1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr1_-_150669604 | 0.07 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr6_+_150690133 | 0.06 |
ENST00000392255.3
ENST00000500320.3 |
IYD
|
iodotyrosine deiodinase |
| chr15_+_74610894 | 0.06 |
ENST00000558821.1
ENST00000268082.4 |
CCDC33
|
coiled-coil domain containing 33 |
| chr3_+_45017722 | 0.06 |
ENST00000265564.7
|
EXOSC7
|
exosome component 7 |
| chr18_+_6834472 | 0.06 |
ENST00000581099.1
ENST00000419673.2 ENST00000531294.1 |
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr16_+_58283814 | 0.06 |
ENST00000443128.2
ENST00000219299.4 |
CCDC113
|
coiled-coil domain containing 113 |
| chr17_+_34312621 | 0.06 |
ENST00000591669.1
|
CTB-186H2.3
|
Uncharacterized protein |
| chr19_-_13227534 | 0.06 |
ENST00000588229.1
ENST00000357720.4 |
TRMT1
|
tRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr18_-_73967160 | 0.06 |
ENST00000579714.1
|
RP11-94B19.7
|
RP11-94B19.7 |
| chr1_-_212004090 | 0.06 |
ENST00000366997.4
|
LPGAT1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr1_+_244227632 | 0.06 |
ENST00000598000.1
|
AL590483.1
|
Uncharacterized protein; cDNA FLJ42623 fis, clone BRACE3015894 |
| chr3_-_69101413 | 0.06 |
ENST00000398559.2
|
TMF1
|
TATA element modulatory factor 1 |
| chr14_-_21567009 | 0.06 |
ENST00000556174.1
ENST00000554478.1 ENST00000553980.1 ENST00000421093.2 |
ZNF219
|
zinc finger protein 219 |
| chrX_+_134847185 | 0.06 |
ENST00000370741.3
ENST00000497301.2 |
CT45A1
|
cancer/testis antigen family 45, member A1 |
| chr12_-_122985067 | 0.06 |
ENST00000540586.1
ENST00000543897.1 |
ZCCHC8
|
zinc finger, CCHC domain containing 8 |
| chr19_-_13227514 | 0.05 |
ENST00000587487.1
ENST00000592814.1 |
TRMT1
|
tRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr9_-_113018835 | 0.05 |
ENST00000374517.5
|
TXN
|
thioredoxin |
| chr19_+_44037546 | 0.05 |
ENST00000601282.1
|
ZNF575
|
zinc finger protein 575 |
| chr19_+_55128576 | 0.05 |
ENST00000396331.1
|
LILRB1
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 1 |
| chr21_+_44394620 | 0.05 |
ENST00000291547.5
|
PKNOX1
|
PBX/knotted 1 homeobox 1 |
| chr7_-_29186008 | 0.05 |
ENST00000396276.3
ENST00000265394.5 |
CPVL
|
carboxypeptidase, vitellogenic-like |
| chr13_-_36705425 | 0.05 |
ENST00000255448.4
ENST00000360631.3 ENST00000379892.4 |
DCLK1
|
doublecortin-like kinase 1 |
| chr2_-_157186630 | 0.05 |
ENST00000406048.2
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr3_-_69101461 | 0.05 |
ENST00000543976.1
|
TMF1
|
TATA element modulatory factor 1 |
| chr3_+_155860751 | 0.05 |
ENST00000471742.1
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chrX_-_134971059 | 0.05 |
ENST00000472834.1
|
CT45A6
|
cancer/testis antigen family 45, member A6 |
| chr18_+_72167096 | 0.05 |
ENST00000324301.8
|
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr12_+_70132632 | 0.05 |
ENST00000378815.6
ENST00000483530.2 ENST00000325555.9 |
RAB3IP
|
RAB3A interacting protein |
| chr14_+_101123580 | 0.05 |
ENST00000556697.1
ENST00000360899.2 ENST00000553623.1 |
LINC00523
|
long intergenic non-protein coding RNA 523 |
| chr3_-_152058779 | 0.05 |
ENST00000408960.3
|
TMEM14E
|
transmembrane protein 14E |
| chrX_-_134971244 | 0.05 |
ENST00000491002.2
ENST00000448053.2 |
CT45A6
|
cancer/testis antigen family 45, member A6 |
| chr3_+_23958632 | 0.05 |
ENST00000412097.1
ENST00000415719.1 ENST00000435882.1 |
RPL15
|
ribosomal protein L15 |
| chr2_+_27760247 | 0.05 |
ENST00000447166.1
|
AC109829.1
|
Uncharacterized protein |
| chr7_-_29152509 | 0.05 |
ENST00000448959.1
|
CPVL
|
carboxypeptidase, vitellogenic-like |
| chr11_-_117102768 | 0.05 |
ENST00000532301.1
|
PCSK7
|
proprotein convertase subtilisin/kexin type 7 |
| chrX_-_134936735 | 0.05 |
ENST00000487941.2
ENST00000434966.2 |
CT45A4
|
cancer/testis antigen family 45, member A4 |
| chr14_-_94970559 | 0.04 |
ENST00000556881.1
|
SERPINA12
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 12 |
| chr3_+_142842128 | 0.04 |
ENST00000483262.1
|
RP11-80H8.4
|
RP11-80H8.4 |
| chr19_-_38183201 | 0.04 |
ENST00000590008.1
ENST00000358582.4 |
ZNF781
|
zinc finger protein 781 |
| chr12_-_120687948 | 0.04 |
ENST00000458477.2
|
PXN
|
paxillin |
| chr7_-_99679324 | 0.04 |
ENST00000292393.5
ENST00000413658.2 ENST00000412947.1 ENST00000441298.1 ENST00000449785.1 ENST00000299667.4 ENST00000424697.1 |
ZNF3
|
zinc finger protein 3 |
| chrX_-_154060946 | 0.04 |
ENST00000369529.1
|
SMIM9
|
small integral membrane protein 9 |
| chr3_+_48413709 | 0.04 |
ENST00000296438.5
ENST00000436231.1 ENST00000445170.1 ENST00000415155.1 |
FBXW12
|
F-box and WD repeat domain containing 12 |
| chr8_-_102216925 | 0.03 |
ENST00000517844.1
|
ZNF706
|
zinc finger protein 706 |
| chr8_-_91095099 | 0.03 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chr2_-_145188137 | 0.03 |
ENST00000440875.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr20_-_4229721 | 0.03 |
ENST00000379453.4
|
ADRA1D
|
adrenoceptor alpha 1D |
| chr18_+_55018044 | 0.03 |
ENST00000324000.3
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr6_+_150690089 | 0.03 |
ENST00000392256.2
|
IYD
|
iodotyrosine deiodinase |
| chr21_+_44394742 | 0.03 |
ENST00000432907.2
|
PKNOX1
|
PBX/knotted 1 homeobox 1 |
| chr2_+_169658928 | 0.03 |
ENST00000317647.7
ENST00000445023.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr3_+_185000729 | 0.03 |
ENST00000448876.1
ENST00000446828.1 ENST00000447637.1 ENST00000424227.1 ENST00000454237.1 |
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr15_-_52944231 | 0.03 |
ENST00000546305.2
|
FAM214A
|
family with sequence similarity 214, member A |
| chr5_-_140700322 | 0.03 |
ENST00000313368.5
|
TAF7
|
TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 55kDa |
| chr6_+_39760129 | 0.03 |
ENST00000274867.4
|
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chr8_-_81787006 | 0.02 |
ENST00000327835.3
|
ZNF704
|
zinc finger protein 704 |
| chr11_+_73358594 | 0.02 |
ENST00000227214.6
ENST00000398494.4 ENST00000543085.1 |
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr16_-_67517716 | 0.02 |
ENST00000290953.2
|
AGRP
|
agouti related protein homolog (mouse) |
| chr3_+_158787098 | 0.02 |
ENST00000397832.2
ENST00000451172.1 ENST00000482126.1 |
IQCJ
|
IQ motif containing J |
| chr7_+_134464414 | 0.02 |
ENST00000361901.2
|
CALD1
|
caldesmon 1 |
| chr12_+_16500037 | 0.02 |
ENST00000536371.1
ENST00000010404.2 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr19_-_7812446 | 0.02 |
ENST00000394173.4
ENST00000301357.8 |
CD209
|
CD209 molecule |
| chr5_+_140535577 | 0.02 |
ENST00000539533.1
|
PCDHB17
|
Protocadherin-psi1; Uncharacterized protein |
| chr4_+_76995855 | 0.02 |
ENST00000355810.4
ENST00000349321.3 |
ART3
|
ADP-ribosyltransferase 3 |
| chr5_+_140480083 | 0.01 |
ENST00000231130.2
|
PCDHB3
|
protocadherin beta 3 |
| chr12_-_10601963 | 0.01 |
ENST00000543893.1
|
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr1_+_111991474 | 0.01 |
ENST00000369722.3
|
ATP5F1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit B1 |
| chr5_+_127039075 | 0.01 |
ENST00000514853.2
|
CTC-228N24.1
|
CTC-228N24.1 |
| chr4_-_1670632 | 0.01 |
ENST00000461064.1
|
FAM53A
|
family with sequence similarity 53, member A |
| chr7_+_134464376 | 0.01 |
ENST00000454108.1
ENST00000361675.2 |
CALD1
|
caldesmon 1 |
| chr1_-_76398077 | 0.01 |
ENST00000284142.6
|
ASB17
|
ankyrin repeat and SOCS box containing 17 |
| chr2_-_238333919 | 0.01 |
ENST00000409162.1
|
AC112721.1
|
Uncharacterized protein |
| chr5_+_140579162 | 0.01 |
ENST00000536699.1
ENST00000354757.3 |
PCDHB11
|
protocadherin beta 11 |
| chr6_+_150690028 | 0.01 |
ENST00000229447.5
ENST00000344419.3 |
IYD
|
iodotyrosine deiodinase |
| chr8_-_107782463 | 0.00 |
ENST00000311955.3
|
ABRA
|
actin-binding Rho activating protein |
| chr15_+_26147507 | 0.00 |
ENST00000383019.2
ENST00000557171.1 |
RP11-1084I9.1
|
RP11-1084I9.1 |
| chr15_-_38519066 | 0.00 |
ENST00000561320.1
ENST00000561161.1 |
RP11-346D14.1
|
RP11-346D14.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of BCL6B
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.1 | 0.4 | GO:1904235 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.1 | 0.3 | GO:0051714 | positive regulation of cytolysis in other organism(GO:0051714) |
| 0.1 | 0.3 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.1 | 0.5 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 0.2 | GO:0034226 | lysine import(GO:0034226) L-lysine import(GO:0061461) L-lysine import into cell(GO:1903410) |
| 0.1 | 0.2 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.1 | 0.4 | GO:2000691 | regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.1 | 0.3 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.1 | 0.2 | GO:1900390 | positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.0 | 0.5 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.2 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.2 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.2 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.2 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.2 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.1 | GO:0010980 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.3 | GO:0045359 | positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 | 0.8 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.1 | GO:1904640 | response to methionine(GO:1904640) |
| 0.0 | 0.1 | GO:0071469 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.3 | GO:0019441 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.0 | 0.1 | GO:0035936 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.0 | 0.2 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.0 | 0.5 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.2 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.1 | GO:2001202 | negative regulation of transforming growth factor-beta secretion(GO:2001202) |
| 0.0 | 0.2 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.1 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:0061590 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.2 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.0 | 0.0 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.0 | 0.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.2 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.5 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.3 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.3 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.1 | 0.3 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.5 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.2 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.1 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.0 | 0.3 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0032396 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) HLA-B specific inhibitory MHC class I receptor activity(GO:0030109) inhibitory MHC class I receptor activity(GO:0032396) |
| 0.0 | 0.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.9 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.2 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.0 | GO:0005499 | vitamin D binding(GO:0005499) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.4 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |