Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
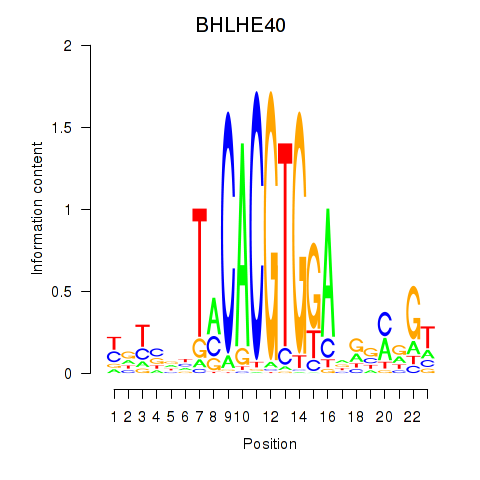
Results for BHLHE40
Z-value: 1.06
Transcription factors associated with BHLHE40
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BHLHE40
|
ENSG00000134107.4 | basic helix-loop-helix family member e40 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BHLHE40 | hg19_v2_chr3_+_5020801_5020952 | -0.88 | 3.3e-07 | Click! |
Activity profile of BHLHE40 motif
Sorted Z-values of BHLHE40 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_32405728 | 2.38 |
ENST00000523079.1
ENST00000338921.4 ENST00000356819.4 ENST00000287845.5 ENST00000341377.5 |
NRG1
|
neuregulin 1 |
| chr21_+_38071430 | 2.31 |
ENST00000290399.6
|
SIM2
|
single-minded family bHLH transcription factor 2 |
| chr14_-_45722360 | 2.12 |
ENST00000451174.1
|
MIS18BP1
|
MIS18 binding protein 1 |
| chr8_+_32405785 | 1.90 |
ENST00000287842.3
|
NRG1
|
neuregulin 1 |
| chr14_-_45722605 | 1.66 |
ENST00000310806.4
|
MIS18BP1
|
MIS18 binding protein 1 |
| chr1_+_203830703 | 1.63 |
ENST00000414487.2
|
SNRPE
|
small nuclear ribonucleoprotein polypeptide E |
| chr1_-_200589859 | 1.59 |
ENST00000367350.4
|
KIF14
|
kinesin family member 14 |
| chrX_+_66764375 | 1.56 |
ENST00000374690.3
|
AR
|
androgen receptor |
| chr16_+_57844549 | 1.50 |
ENST00000564282.1
|
CTD-2600O9.1
|
uncharacterized protein LOC388282 |
| chr19_-_16045619 | 1.35 |
ENST00000402119.4
|
CYP4F11
|
cytochrome P450, family 4, subfamily F, polypeptide 11 |
| chr19_-_16045665 | 1.34 |
ENST00000248041.8
|
CYP4F11
|
cytochrome P450, family 4, subfamily F, polypeptide 11 |
| chr17_-_53046058 | 1.27 |
ENST00000571584.1
ENST00000299335.3 |
COX11
|
cytochrome c oxidase assembly homolog 11 (yeast) |
| chr2_+_103236004 | 1.26 |
ENST00000233969.2
|
SLC9A2
|
solute carrier family 9, subfamily A (NHE2, cation proton antiporter 2), member 2 |
| chr17_+_7982800 | 1.17 |
ENST00000399413.3
|
AC129492.6
|
AC129492.6 |
| chr12_+_65004292 | 1.14 |
ENST00000542104.1
ENST00000336061.2 |
RASSF3
|
Ras association (RalGDS/AF-6) domain family member 3 |
| chr9_+_131901710 | 1.11 |
ENST00000524946.2
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr15_+_77713299 | 1.11 |
ENST00000559099.1
|
HMG20A
|
high mobility group 20A |
| chr17_-_74733404 | 1.10 |
ENST00000508921.3
ENST00000583836.1 ENST00000358156.6 ENST00000392485.2 ENST00000359995.5 |
SRSF2
|
serine/arginine-rich splicing factor 2 |
| chr2_-_136743436 | 1.03 |
ENST00000441323.1
ENST00000449218.1 |
DARS
|
aspartyl-tRNA synthetase |
| chr6_+_151561085 | 1.01 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr2_-_240230890 | 1.01 |
ENST00000446876.1
|
HDAC4
|
histone deacetylase 4 |
| chr17_+_33307503 | 0.98 |
ENST00000378526.4
ENST00000585941.1 ENST00000262327.5 ENST00000592690.1 ENST00000585740.1 |
LIG3
|
ligase III, DNA, ATP-dependent |
| chr9_+_131901661 | 0.95 |
ENST00000423100.1
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr22_-_43411106 | 0.95 |
ENST00000453643.1
ENST00000263246.3 ENST00000337959.4 |
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr2_-_179343226 | 0.95 |
ENST00000434643.2
|
FKBP7
|
FK506 binding protein 7 |
| chr17_+_53046096 | 0.94 |
ENST00000376352.2
ENST00000299341.4 ENST00000405898.1 ENST00000434978.2 ENST00000398391.2 |
STXBP4
|
syntaxin binding protein 4 |
| chr13_+_35516390 | 0.94 |
ENST00000540320.1
ENST00000400445.3 ENST00000310336.4 |
NBEA
|
neurobeachin |
| chr19_+_54058073 | 0.92 |
ENST00000505949.1
ENST00000513265.1 |
ZNF331
|
zinc finger protein 331 |
| chr13_+_50656307 | 0.91 |
ENST00000378180.4
|
DLEU1
|
deleted in lymphocytic leukemia 1 (non-protein coding) |
| chr1_+_213123976 | 0.90 |
ENST00000366965.2
ENST00000366967.2 |
VASH2
|
vasohibin 2 |
| chr6_-_36842784 | 0.89 |
ENST00000373699.5
|
PPIL1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr1_+_173793777 | 0.88 |
ENST00000239457.5
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr2_+_183989157 | 0.88 |
ENST00000541912.1
|
NUP35
|
nucleoporin 35kDa |
| chr2_+_190649107 | 0.87 |
ENST00000441310.2
ENST00000409985.1 ENST00000446877.1 ENST00000418224.3 ENST00000409823.3 ENST00000374826.4 ENST00000424766.1 ENST00000447232.2 ENST00000432292.3 |
PMS1
|
PMS1 postmeiotic segregation increased 1 (S. cerevisiae) |
| chr9_+_131902283 | 0.86 |
ENST00000436883.1
ENST00000414510.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr2_-_112642267 | 0.85 |
ENST00000341068.3
|
ANAPC1
|
anaphase promoting complex subunit 1 |
| chr17_+_74732889 | 0.85 |
ENST00000591864.1
|
MFSD11
|
major facilitator superfamily domain containing 11 |
| chr6_-_136871957 | 0.84 |
ENST00000354570.3
|
MAP7
|
microtubule-associated protein 7 |
| chr2_+_183989083 | 0.84 |
ENST00000295119.4
|
NUP35
|
nucleoporin 35kDa |
| chr16_+_66461175 | 0.83 |
ENST00000536005.2
ENST00000299694.8 ENST00000561796.1 |
BEAN1
|
brain expressed, associated with NEDD4, 1 |
| chr8_+_99129513 | 0.82 |
ENST00000522319.1
ENST00000401707.2 |
POP1
|
processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae) |
| chrX_+_54556633 | 0.81 |
ENST00000336470.4
ENST00000360845.2 |
GNL3L
|
guanine nucleotide binding protein-like 3 (nucleolar)-like |
| chr21_+_33784670 | 0.80 |
ENST00000300255.2
|
EVA1C
|
eva-1 homolog C (C. elegans) |
| chr2_-_241497390 | 0.79 |
ENST00000272972.3
ENST00000401804.1 ENST00000361678.4 ENST00000405523.3 |
ANKMY1
|
ankyrin repeat and MYND domain containing 1 |
| chr15_+_77713222 | 0.78 |
ENST00000558176.1
|
HMG20A
|
high mobility group 20A |
| chr4_-_57301748 | 0.78 |
ENST00000264220.2
|
PPAT
|
phosphoribosyl pyrophosphate amidotransferase |
| chr8_-_99129338 | 0.75 |
ENST00000520507.1
|
HRSP12
|
heat-responsive protein 12 |
| chr6_+_36410762 | 0.75 |
ENST00000483557.1
ENST00000498267.1 ENST00000544295.1 ENST00000449081.2 ENST00000536244.1 ENST00000460983.1 |
KCTD20
|
potassium channel tetramerization domain containing 20 |
| chr17_+_72428266 | 0.74 |
ENST00000582473.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr8_+_55047763 | 0.71 |
ENST00000260102.4
ENST00000519831.1 |
MRPL15
|
mitochondrial ribosomal protein L15 |
| chr1_+_213123915 | 0.70 |
ENST00000366968.4
ENST00000490792.1 |
VASH2
|
vasohibin 2 |
| chr5_-_89770171 | 0.70 |
ENST00000514906.1
|
MBLAC2
|
metallo-beta-lactamase domain containing 2 |
| chr11_-_57282349 | 0.70 |
ENST00000528450.1
|
SLC43A1
|
solute carrier family 43 (amino acid system L transporter), member 1 |
| chr2_-_47143160 | 0.70 |
ENST00000409800.1
ENST00000409218.1 |
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr2_+_207630081 | 0.70 |
ENST00000236980.6
ENST00000418289.1 ENST00000402774.3 ENST00000403094.3 |
FASTKD2
|
FAST kinase domains 2 |
| chr1_+_213123862 | 0.70 |
ENST00000366966.2
ENST00000366964.3 |
VASH2
|
vasohibin 2 |
| chr8_-_48872686 | 0.68 |
ENST00000314191.2
ENST00000338368.3 |
PRKDC
|
protein kinase, DNA-activated, catalytic polypeptide |
| chr4_-_76861392 | 0.68 |
ENST00000505594.1
|
NAAA
|
N-acylethanolamine acid amidase |
| chr16_-_19729453 | 0.67 |
ENST00000564480.1
|
KNOP1
|
lysine-rich nucleolar protein 1 |
| chr12_-_25101920 | 0.66 |
ENST00000539780.1
ENST00000546285.1 ENST00000342945.5 |
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr19_-_40786733 | 0.66 |
ENST00000486368.2
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr3_-_51533966 | 0.66 |
ENST00000504652.1
|
VPRBP
|
Vpr (HIV-1) binding protein |
| chr10_-_35379241 | 0.65 |
ENST00000374748.1
ENST00000374749.3 |
CUL2
|
cullin 2 |
| chr11_-_3147835 | 0.65 |
ENST00000525498.1
|
OSBPL5
|
oxysterol binding protein-like 5 |
| chr2_-_47142884 | 0.65 |
ENST00000409105.1
ENST00000409973.1 ENST00000409913.1 ENST00000319466.4 |
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr10_-_35379524 | 0.61 |
ENST00000374751.3
ENST00000374742.1 ENST00000602371.1 |
CUL2
|
cullin 2 |
| chr9_-_139258235 | 0.60 |
ENST00000371738.3
|
DNLZ
|
DNL-type zinc finger |
| chr5_+_89770664 | 0.58 |
ENST00000503973.1
ENST00000399107.1 |
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr1_+_210001309 | 0.57 |
ENST00000491415.2
|
DIEXF
|
digestive organ expansion factor homolog (zebrafish) |
| chr7_-_138720763 | 0.56 |
ENST00000275766.1
|
ZC3HAV1L
|
zinc finger CCCH-type, antiviral 1-like |
| chrX_+_23685653 | 0.56 |
ENST00000379331.3
|
PRDX4
|
peroxiredoxin 4 |
| chr2_-_241497374 | 0.56 |
ENST00000373318.2
ENST00000406958.1 ENST00000391987.1 ENST00000373320.4 |
ANKMY1
|
ankyrin repeat and MYND domain containing 1 |
| chr2_-_179343268 | 0.55 |
ENST00000424785.2
|
FKBP7
|
FK506 binding protein 7 |
| chr5_+_36152091 | 0.55 |
ENST00000274254.5
|
SKP2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr11_+_60609537 | 0.54 |
ENST00000227520.5
|
CCDC86
|
coiled-coil domain containing 86 |
| chr5_+_421030 | 0.54 |
ENST00000506456.1
|
AHRR
|
aryl-hydrocarbon receptor repressor |
| chr17_-_17109579 | 0.54 |
ENST00000321560.3
|
PLD6
|
phospholipase D family, member 6 |
| chr10_+_49514698 | 0.54 |
ENST00000432379.1
ENST00000429041.1 ENST00000374189.1 |
MAPK8
|
mitogen-activated protein kinase 8 |
| chr14_+_73393040 | 0.53 |
ENST00000358377.2
ENST00000353777.3 ENST00000394234.2 ENST00000509153.1 ENST00000555042.1 |
DCAF4
|
DDB1 and CUL4 associated factor 4 |
| chr4_-_15683118 | 0.51 |
ENST00000507899.1
ENST00000510802.1 |
FBXL5
|
F-box and leucine-rich repeat protein 5 |
| chr12_-_58165870 | 0.50 |
ENST00000257848.7
|
METTL1
|
methyltransferase like 1 |
| chr2_+_201676908 | 0.48 |
ENST00000409226.1
ENST00000452790.2 |
BZW1
|
basic leucine zipper and W2 domains 1 |
| chr12_-_6677422 | 0.48 |
ENST00000382421.3
ENST00000545200.1 ENST00000399466.2 ENST00000536124.1 ENST00000540228.1 ENST00000542867.1 ENST00000545492.1 ENST00000322166.5 ENST00000545915.1 |
NOP2
|
NOP2 nucleolar protein |
| chr11_-_61560053 | 0.48 |
ENST00000537328.1
|
TMEM258
|
transmembrane protein 258 |
| chr12_+_93861282 | 0.47 |
ENST00000552217.1
ENST00000393128.4 ENST00000547098.1 |
MRPL42
|
mitochondrial ribosomal protein L42 |
| chr5_-_89770582 | 0.46 |
ENST00000316610.6
|
MBLAC2
|
metallo-beta-lactamase domain containing 2 |
| chr8_+_125985531 | 0.46 |
ENST00000319286.5
|
ZNF572
|
zinc finger protein 572 |
| chr12_-_25102252 | 0.44 |
ENST00000261192.7
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr17_+_72428218 | 0.44 |
ENST00000392628.2
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr11_-_65626753 | 0.43 |
ENST00000526975.1
ENST00000531413.1 |
CFL1
|
cofilin 1 (non-muscle) |
| chrX_-_16887963 | 0.42 |
ENST00000380084.4
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr2_-_10587897 | 0.42 |
ENST00000405333.1
ENST00000443218.1 |
ODC1
|
ornithine decarboxylase 1 |
| chr2_-_10588630 | 0.42 |
ENST00000234111.4
|
ODC1
|
ornithine decarboxylase 1 |
| chr1_+_231376941 | 0.41 |
ENST00000436239.1
ENST00000366647.4 ENST00000366646.3 ENST00000416000.1 |
GNPAT
|
glyceronephosphate O-acyltransferase |
| chr1_-_43638168 | 0.40 |
ENST00000431635.2
|
EBNA1BP2
|
EBNA1 binding protein 2 |
| chr7_-_107642348 | 0.38 |
ENST00000393561.1
|
LAMB1
|
laminin, beta 1 |
| chr11_+_6624955 | 0.37 |
ENST00000299421.4
ENST00000537806.1 |
ILK
|
integrin-linked kinase |
| chr5_-_71616043 | 0.36 |
ENST00000508863.2
ENST00000522095.1 ENST00000513900.1 ENST00000515404.1 ENST00000457646.4 ENST00000261413.5 |
MRPS27
|
mitochondrial ribosomal protein S27 |
| chr17_-_61850894 | 0.35 |
ENST00000403162.3
ENST00000582252.1 ENST00000225726.5 |
CCDC47
|
coiled-coil domain containing 47 |
| chr5_+_172571445 | 0.35 |
ENST00000231668.9
ENST00000351486.5 ENST00000352523.6 ENST00000393770.4 |
BNIP1
|
BCL2/adenovirus E1B 19kDa interacting protein 1 |
| chr5_+_36152179 | 0.35 |
ENST00000508514.1
ENST00000513151.1 ENST00000546211.1 |
SKP2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr12_+_93861264 | 0.34 |
ENST00000549982.1
ENST00000361630.2 |
MRPL42
|
mitochondrial ribosomal protein L42 |
| chr18_+_55712915 | 0.34 |
ENST00000592846.1
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr16_+_29911864 | 0.33 |
ENST00000308748.5
|
ASPHD1
|
aspartate beta-hydroxylase domain containing 1 |
| chr2_-_136743039 | 0.32 |
ENST00000537273.1
|
DARS
|
aspartyl-tRNA synthetase |
| chrX_+_129040094 | 0.32 |
ENST00000425117.2
|
UTP14A
|
UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) |
| chr5_+_89770696 | 0.32 |
ENST00000504930.1
ENST00000514483.1 |
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr2_+_74154032 | 0.31 |
ENST00000356837.6
|
DGUOK
|
deoxyguanosine kinase |
| chr3_-_98312548 | 0.30 |
ENST00000264193.2
|
CPOX
|
coproporphyrinogen oxidase |
| chr11_+_6624970 | 0.30 |
ENST00000420936.2
ENST00000528995.1 |
ILK
|
integrin-linked kinase |
| chr14_+_93389425 | 0.30 |
ENST00000216492.5
ENST00000334654.4 |
CHGA
|
chromogranin A (parathyroid secretory protein 1) |
| chrX_+_129040122 | 0.30 |
ENST00000394422.3
ENST00000371051.5 |
UTP14A
|
UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) |
| chr20_-_42939782 | 0.29 |
ENST00000396825.3
|
FITM2
|
fat storage-inducing transmembrane protein 2 |
| chr10_+_101491968 | 0.28 |
ENST00000370476.5
ENST00000370472.4 |
CUTC
|
cutC copper transporter |
| chr1_-_43637915 | 0.28 |
ENST00000236051.2
|
EBNA1BP2
|
EBNA1 binding protein 2 |
| chr7_-_229557 | 0.28 |
ENST00000514988.1
|
AC145676.2
|
Uncharacterized protein |
| chr7_-_37024665 | 0.28 |
ENST00000396040.2
|
ELMO1
|
engulfment and cell motility 1 |
| chr19_-_55652290 | 0.27 |
ENST00000589745.1
|
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr17_-_10600818 | 0.27 |
ENST00000577427.1
ENST00000255390.5 |
SCO1
|
SCO1 cytochrome c oxidase assembly protein |
| chr2_-_190649023 | 0.26 |
ENST00000409519.1
ENST00000458355.1 |
ORMDL1
|
ORM1-like 1 (S. cerevisiae) |
| chr12_-_55042140 | 0.26 |
ENST00000293371.6
ENST00000456047.2 |
DCD
|
dermcidin |
| chrX_-_47518498 | 0.26 |
ENST00000335890.2
|
UXT
|
ubiquitously-expressed, prefoldin-like chaperone |
| chrX_-_47518527 | 0.25 |
ENST00000333119.3
|
UXT
|
ubiquitously-expressed, prefoldin-like chaperone |
| chr15_-_35088340 | 0.25 |
ENST00000290378.4
|
ACTC1
|
actin, alpha, cardiac muscle 1 |
| chr15_+_68924327 | 0.24 |
ENST00000543950.1
|
CORO2B
|
coronin, actin binding protein, 2B |
| chr11_+_6625046 | 0.24 |
ENST00000396751.2
|
ILK
|
integrin-linked kinase |
| chr5_-_36152031 | 0.24 |
ENST00000296603.4
|
LMBRD2
|
LMBR1 domain containing 2 |
| chr3_+_99536663 | 0.24 |
ENST00000421999.2
ENST00000463526.1 |
CMSS1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr4_+_57301896 | 0.23 |
ENST00000514888.1
ENST00000264221.2 ENST00000505164.1 |
PAICS
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr22_-_30953587 | 0.23 |
ENST00000453479.1
|
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr17_+_36908984 | 0.22 |
ENST00000225426.4
ENST00000579088.1 |
PSMB3
|
proteasome (prosome, macropain) subunit, beta type, 3 |
| chr9_-_100881466 | 0.21 |
ENST00000341469.2
ENST00000342043.3 ENST00000375098.3 |
TRIM14
|
tripartite motif containing 14 |
| chr22_+_48885272 | 0.21 |
ENST00000402357.1
|
FAM19A5
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5 |
| chr3_+_63638372 | 0.20 |
ENST00000496807.1
|
SNTN
|
sentan, cilia apical structure protein |
| chr1_+_118472343 | 0.19 |
ENST00000369441.3
ENST00000349139.5 |
WDR3
|
WD repeat domain 3 |
| chr16_+_19729586 | 0.18 |
ENST00000564186.1
ENST00000541926.1 ENST00000433597.2 |
IQCK
|
IQ motif containing K |
| chr5_-_114880533 | 0.18 |
ENST00000274457.3
|
FEM1C
|
fem-1 homolog c (C. elegans) |
| chr19_+_56154913 | 0.18 |
ENST00000270451.5
ENST00000588537.1 |
ZNF581
|
zinc finger protein 581 |
| chr19_+_45909893 | 0.17 |
ENST00000592852.1
ENST00000589804.1 ENST00000590794.1 |
CD3EAP
|
CD3e molecule, epsilon associated protein |
| chr20_+_23331373 | 0.17 |
ENST00000254998.2
|
NXT1
|
NTF2-like export factor 1 |
| chr1_+_9648921 | 0.16 |
ENST00000377376.4
ENST00000340305.5 ENST00000340381.6 |
TMEM201
|
transmembrane protein 201 |
| chr6_+_5261225 | 0.16 |
ENST00000324331.6
|
FARS2
|
phenylalanyl-tRNA synthetase 2, mitochondrial |
| chr9_+_130469257 | 0.16 |
ENST00000373295.2
|
C9orf117
|
chromosome 9 open reading frame 117 |
| chr2_+_105654441 | 0.16 |
ENST00000258455.3
|
MRPS9
|
mitochondrial ribosomal protein S9 |
| chr17_-_8661860 | 0.16 |
ENST00000328794.6
|
SPDYE4
|
speedy/RINGO cell cycle regulator family member E4 |
| chr1_-_171621815 | 0.15 |
ENST00000037502.6
|
MYOC
|
myocilin, trabecular meshwork inducible glucocorticoid response |
| chr5_+_36152163 | 0.15 |
ENST00000274255.6
|
SKP2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr17_+_2699697 | 0.14 |
ENST00000254695.8
ENST00000366401.4 ENST00000542807.1 |
RAP1GAP2
|
RAP1 GTPase activating protein 2 |
| chr15_+_58724184 | 0.14 |
ENST00000433326.2
|
LIPC
|
lipase, hepatic |
| chr9_-_95640218 | 0.14 |
ENST00000395506.3
ENST00000375495.3 ENST00000332591.6 |
ZNF484
|
zinc finger protein 484 |
| chr4_-_100009856 | 0.13 |
ENST00000296412.8
|
ADH5
|
alcohol dehydrogenase 5 (class III), chi polypeptide |
| chr6_+_44215603 | 0.13 |
ENST00000371554.1
|
HSP90AB1
|
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chr19_-_3786253 | 0.12 |
ENST00000585778.1
|
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr4_-_85418103 | 0.12 |
ENST00000515820.2
|
NKX6-1
|
NK6 homeobox 1 |
| chr7_+_141463897 | 0.12 |
ENST00000247879.2
|
TAS2R3
|
taste receptor, type 2, member 3 |
| chr15_-_74988281 | 0.11 |
ENST00000566828.1
ENST00000563009.1 ENST00000568176.1 ENST00000566243.1 ENST00000566219.1 ENST00000426797.3 ENST00000566119.1 ENST00000315127.4 |
EDC3
|
enhancer of mRNA decapping 3 |
| chr8_-_99129384 | 0.11 |
ENST00000521560.1
ENST00000254878.3 |
HRSP12
|
heat-responsive protein 12 |
| chr1_+_119957554 | 0.11 |
ENST00000543831.1
ENST00000433745.1 ENST00000369416.3 |
HSD3B2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr19_+_51774540 | 0.11 |
ENST00000600813.1
|
CTD-3187F8.11
|
CTD-3187F8.11 |
| chr1_-_207095324 | 0.11 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr9_-_99540328 | 0.10 |
ENST00000223428.4
ENST00000375231.1 ENST00000374641.3 |
ZNF510
|
zinc finger protein 510 |
| chr7_-_100493744 | 0.10 |
ENST00000428317.1
ENST00000441605.1 |
ACHE
|
acetylcholinesterase (Yt blood group) |
| chr22_+_22681656 | 0.10 |
ENST00000390291.2
|
IGLV1-50
|
immunoglobulin lambda variable 1-50 (non-functional) |
| chr6_-_109804412 | 0.09 |
ENST00000230122.3
|
ZBTB24
|
zinc finger and BTB domain containing 24 |
| chr11_-_65626797 | 0.09 |
ENST00000525451.2
|
CFL1
|
cofilin 1 (non-muscle) |
| chr10_-_49459800 | 0.09 |
ENST00000305531.3
|
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr17_+_61851157 | 0.09 |
ENST00000578681.1
ENST00000583590.1 |
DDX42
|
DEAD (Asp-Glu-Ala-Asp) box helicase 42 |
| chr5_+_179078298 | 0.08 |
ENST00000418535.2
ENST00000425471.1 |
AC136604.1
|
Uncharacterized protein |
| chr16_-_3149229 | 0.08 |
ENST00000572431.1
ENST00000572548.1 |
ZSCAN10
|
zinc finger and SCAN domain containing 10 |
| chr2_+_74153953 | 0.07 |
ENST00000264093.4
ENST00000348222.1 |
DGUOK
|
deoxyguanosine kinase |
| chr7_+_5919458 | 0.07 |
ENST00000416608.1
|
OCM
|
oncomodulin |
| chr6_+_13615554 | 0.07 |
ENST00000451315.2
|
NOL7
|
nucleolar protein 7, 27kDa |
| chr17_+_61851504 | 0.06 |
ENST00000359353.5
ENST00000389924.2 |
DDX42
|
DEAD (Asp-Glu-Ala-Asp) box helicase 42 |
| chr5_+_6766004 | 0.06 |
ENST00000506093.1
|
RP11-332J15.3
|
RP11-332J15.3 |
| chr1_-_26700943 | 0.05 |
ENST00000416125.1
|
ZNF683
|
zinc finger protein 683 |
| chr11_+_73019282 | 0.05 |
ENST00000263674.3
|
ARHGEF17
|
Rho guanine nucleotide exchange factor (GEF) 17 |
| chr1_-_231376867 | 0.05 |
ENST00000366649.2
ENST00000318906.2 ENST00000366651.3 |
C1orf131
|
chromosome 1 open reading frame 131 |
| chr1_-_86174065 | 0.05 |
ENST00000370574.3
ENST00000431532.2 |
ZNHIT6
|
zinc finger, HIT-type containing 6 |
| chr2_-_136743169 | 0.04 |
ENST00000264161.4
|
DARS
|
aspartyl-tRNA synthetase |
| chr3_+_111578583 | 0.04 |
ENST00000478922.1
ENST00000477695.1 |
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr6_-_154677866 | 0.04 |
ENST00000367220.4
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr1_+_43637996 | 0.03 |
ENST00000528956.1
ENST00000529956.1 |
WDR65
|
WD repeat domain 65 |
| chr5_+_149737202 | 0.03 |
ENST00000451292.1
ENST00000377797.3 ENST00000445265.2 ENST00000323668.7 ENST00000439160.2 ENST00000394269.3 ENST00000427724.2 ENST00000504761.2 ENST00000513346.1 ENST00000515516.1 |
TCOF1
|
Treacher Collins-Franceschetti syndrome 1 |
| chr14_+_101359265 | 0.03 |
ENST00000599197.1
|
AL117190.3
|
Esophagus cancer-related gene-2 interaction susceptibility protein; Uncharacterized protein |
| chr1_+_20465805 | 0.02 |
ENST00000375102.3
|
PLA2G2F
|
phospholipase A2, group IIF |
| chr7_-_151574191 | 0.02 |
ENST00000287878.4
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr4_+_41983713 | 0.02 |
ENST00000333141.5
|
DCAF4L1
|
DDB1 and CUL4 associated factor 4-like 1 |
| chr17_-_7193711 | 0.01 |
ENST00000571464.1
|
YBX2
|
Y box binding protein 2 |
| chr7_-_155326532 | 0.01 |
ENST00000406197.1
ENST00000321736.5 |
CNPY1
|
canopy FGF signaling regulator 1 |
| chr7_-_73133959 | 0.01 |
ENST00000395155.3
ENST00000395154.3 ENST00000222812.3 ENST00000395156.3 |
STX1A
|
syntaxin 1A (brain) |
| chr13_+_111972980 | 0.00 |
ENST00000283547.1
|
TEX29
|
testis expressed 29 |
| chr8_+_109455845 | 0.00 |
ENST00000220853.3
|
EMC2
|
ER membrane protein complex subunit 2 |
| chr8_+_109455830 | 0.00 |
ENST00000524143.1
|
EMC2
|
ER membrane protein complex subunit 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of BHLHE40
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.7 | GO:0042377 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.5 | 1.6 | GO:0045720 | negative regulation of integrin biosynthetic process(GO:0045720) |
| 0.5 | 4.3 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.3 | 1.4 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.3 | 1.6 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.2 | 0.9 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.2 | 1.3 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.2 | 1.0 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.2 | 1.0 | GO:0006288 | base-excision repair, DNA ligation(GO:0006288) |
| 0.2 | 1.1 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.2 | 0.5 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.2 | 0.2 | GO:0038129 | ERBB3 signaling pathway(GO:0038129) |
| 0.1 | 0.7 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 0.8 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.8 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.1 | 0.7 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 0.8 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.5 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.1 | 2.7 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 0.3 | GO:1900738 | dense core granule biogenesis(GO:0061110) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 | 4.2 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 0.5 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 0.4 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.1 | 0.7 | GO:0071486 | cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.1 | 0.4 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 1.0 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.1 | 0.4 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.1 | 0.5 | GO:2000782 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.1 | 1.0 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 2.3 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.1 | 0.5 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 1.3 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 1.0 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.1 | 0.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.9 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.1 | 1.7 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.1 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.0 | 1.1 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.3 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.2 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.0 | 2.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.0 | 0.3 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.9 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 1.6 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.0 | 0.3 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.3 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 1.5 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.3 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.9 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 2.3 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 0.7 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 0.2 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 1.9 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.5 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.9 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:0002325 | natural killer cell differentiation involved in immune response(GO:0002325) regulation of natural killer cell differentiation involved in immune response(GO:0032826) regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.0 | 0.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 1.5 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.0 | 1.3 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.5 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.5 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.7 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.8 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.2 | 1.6 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.2 | 1.3 | GO:0030891 | VCB complex(GO:0030891) |
| 0.2 | 1.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.1 | 0.4 | GO:0043257 | laminin-8 complex(GO:0043257) |
| 0.1 | 0.8 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.7 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 0.9 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 0.5 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.1 | 1.7 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 3.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.5 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 1.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.3 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 1.4 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.3 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 0.9 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.6 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 1.0 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.8 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.5 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 1.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.9 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 1.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.2 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 1.0 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.3 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.4 | 4.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.3 | 1.6 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.2 | 1.1 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.2 | 0.5 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.2 | 1.6 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.1 | 0.5 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.1 | 1.0 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.1 | 1.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.6 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 1.0 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.5 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.1 | 0.6 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.4 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 1.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.5 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 2.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.8 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 1.5 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.4 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 1.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.7 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.1 | GO:0002134 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.9 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.5 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 1.0 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.3 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.3 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.3 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.2 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 1.1 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 1.1 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 1.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.8 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.7 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.0 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.6 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.7 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.5 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 1.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.6 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.7 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 2.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.9 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.6 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 4.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.0 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 1.1 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 2.4 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.0 | 1.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 1.7 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.8 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.5 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 1.1 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.9 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 1.7 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.6 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 1.3 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.7 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |