Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
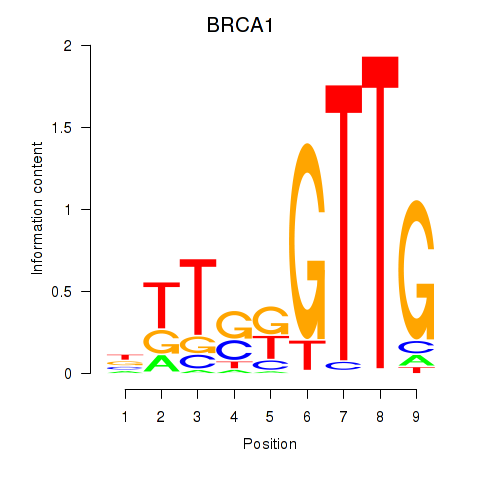
Results for BRCA1
Z-value: 0.34
Transcription factors associated with BRCA1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BRCA1
|
ENSG00000012048.15 | BRCA1 DNA repair associated |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BRCA1 | hg19_v2_chr17_-_41277467_41277510 | -0.35 | 1.4e-01 | Click! |
Activity profile of BRCA1 motif
Sorted Z-values of BRCA1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_117947782 | 1.97 |
ENST00000522307.1
ENST00000523251.1 ENST00000437212.3 ENST00000522824.1 ENST00000522151.1 |
TMPRSS4
|
transmembrane protease, serine 4 |
| chr3_-_176914191 | 1.34 |
ENST00000437738.1
ENST00000424913.1 ENST00000443315.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr14_+_21510385 | 1.26 |
ENST00000298690.4
|
RNASE7
|
ribonuclease, RNase A family, 7 |
| chr11_+_117947724 | 1.15 |
ENST00000534111.1
|
TMPRSS4
|
transmembrane protease, serine 4 |
| chr14_-_69261310 | 0.99 |
ENST00000336440.3
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr17_+_36584662 | 0.84 |
ENST00000431231.2
ENST00000437668.3 |
ARHGAP23
|
Rho GTPase activating protein 23 |
| chr17_+_74372662 | 0.68 |
ENST00000591651.1
ENST00000545180.1 |
SPHK1
|
sphingosine kinase 1 |
| chr4_-_48782259 | 0.65 |
ENST00000507711.1
ENST00000358350.4 ENST00000537810.1 ENST00000264319.7 |
FRYL
|
FRY-like |
| chr3_-_176914238 | 0.64 |
ENST00000430069.1
ENST00000428970.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr7_+_44646177 | 0.53 |
ENST00000443864.2
ENST00000447398.1 ENST00000449767.1 ENST00000419661.1 |
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr11_-_13484713 | 0.51 |
ENST00000526841.1
ENST00000529708.1 ENST00000278174.5 ENST00000528120.1 |
BTBD10
|
BTB (POZ) domain containing 10 |
| chr5_+_145583107 | 0.51 |
ENST00000506502.1
|
RBM27
|
RNA binding motif protein 27 |
| chr18_+_55888767 | 0.50 |
ENST00000431212.2
ENST00000586268.1 ENST00000587190.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr18_-_23671139 | 0.49 |
ENST00000579061.1
ENST00000542420.2 |
SS18
|
synovial sarcoma translocation, chromosome 18 |
| chr10_+_122610853 | 0.48 |
ENST00000604585.1
|
WDR11
|
WD repeat domain 11 |
| chr12_-_122879969 | 0.47 |
ENST00000540304.1
|
CLIP1
|
CAP-GLY domain containing linker protein 1 |
| chr10_-_75385711 | 0.44 |
ENST00000433394.1
|
USP54
|
ubiquitin specific peptidase 54 |
| chr1_-_115301235 | 0.41 |
ENST00000525878.1
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr10_+_28822636 | 0.40 |
ENST00000442148.1
ENST00000448193.1 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chr13_-_50018241 | 0.39 |
ENST00000409308.1
|
CAB39L
|
calcium binding protein 39-like |
| chr5_-_68665084 | 0.38 |
ENST00000509462.1
|
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chr12_-_49418407 | 0.37 |
ENST00000526209.1
|
KMT2D
|
lysine (K)-specific methyltransferase 2D |
| chr7_+_44646162 | 0.36 |
ENST00000439616.2
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr13_-_50018140 | 0.35 |
ENST00000410043.1
ENST00000347776.5 |
CAB39L
|
calcium binding protein 39-like |
| chr1_-_160232312 | 0.34 |
ENST00000440682.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr8_-_116681123 | 0.34 |
ENST00000519674.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr14_+_32030582 | 0.34 |
ENST00000550649.1
ENST00000281081.7 |
NUBPL
|
nucleotide binding protein-like |
| chr5_+_145583156 | 0.31 |
ENST00000265271.5
|
RBM27
|
RNA binding motif protein 27 |
| chr5_-_124082279 | 0.31 |
ENST00000513986.1
|
ZNF608
|
zinc finger protein 608 |
| chr2_+_203879568 | 0.31 |
ENST00000449802.1
|
NBEAL1
|
neurobeachin-like 1 |
| chr8_-_116681221 | 0.31 |
ENST00000395715.3
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr19_-_37663572 | 0.28 |
ENST00000588354.1
ENST00000292841.5 ENST00000355533.2 ENST00000356958.4 |
ZNF585A
|
zinc finger protein 585A |
| chr12_-_67072714 | 0.28 |
ENST00000545666.1
ENST00000398016.3 ENST00000359742.4 ENST00000286445.7 ENST00000538211.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr1_+_70687079 | 0.27 |
ENST00000395136.2
|
SRSF11
|
serine/arginine-rich splicing factor 11 |
| chr19_-_37663332 | 0.24 |
ENST00000392157.2
|
ZNF585A
|
zinc finger protein 585A |
| chr2_+_223725652 | 0.24 |
ENST00000357430.3
ENST00000392066.3 |
ACSL3
|
acyl-CoA synthetase long-chain family member 3 |
| chr1_-_227505289 | 0.24 |
ENST00000366765.3
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like) |
| chr6_-_42981651 | 0.23 |
ENST00000244711.3
|
MEA1
|
male-enhanced antigen 1 |
| chr8_+_22022800 | 0.22 |
ENST00000397814.3
|
BMP1
|
bone morphogenetic protein 1 |
| chr14_+_103058948 | 0.21 |
ENST00000262241.6
|
RCOR1
|
REST corepressor 1 |
| chr20_+_9049682 | 0.21 |
ENST00000334005.3
ENST00000378473.3 |
PLCB4
|
phospholipase C, beta 4 |
| chr8_+_22022653 | 0.21 |
ENST00000354870.5
ENST00000397816.3 ENST00000306349.8 |
BMP1
|
bone morphogenetic protein 1 |
| chr5_-_68664989 | 0.20 |
ENST00000508954.1
|
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chr3_+_185303962 | 0.20 |
ENST00000296257.5
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr12_-_49449107 | 0.20 |
ENST00000301067.7
|
KMT2D
|
lysine (K)-specific methyltransferase 2D |
| chr2_-_134326009 | 0.20 |
ENST00000409261.1
ENST00000409213.1 |
NCKAP5
|
NCK-associated protein 5 |
| chr14_-_75179774 | 0.19 |
ENST00000555249.1
ENST00000556202.1 ENST00000356357.4 ENST00000338772.5 |
AREL1
AC007956.1
|
apoptosis resistant E3 ubiquitin protein ligase 1 Full-length cDNA 5-PRIME end of clone CS0CAP004YO05 of Thymus of Homo sapiens (human); Uncharacterized protein |
| chr20_+_52105495 | 0.18 |
ENST00000439873.2
|
AL354993.1
|
Cell growth-inhibiting protein 7; HCG1784586; Uncharacterized protein |
| chr10_-_61900762 | 0.18 |
ENST00000355288.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr10_+_102295616 | 0.18 |
ENST00000299163.6
|
HIF1AN
|
hypoxia inducible factor 1, alpha subunit inhibitor |
| chr13_+_27825706 | 0.17 |
ENST00000272274.4
ENST00000319826.4 ENST00000326092.4 |
RPL21
|
ribosomal protein L21 |
| chr5_+_140079919 | 0.16 |
ENST00000274712.3
|
ZMAT2
|
zinc finger, matrin-type 2 |
| chr13_-_103046837 | 0.16 |
ENST00000607251.1
|
FGF14-IT1
|
FGF14 intronic transcript 1 (non-protein coding) |
| chr5_-_68665469 | 0.15 |
ENST00000217893.5
|
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chr12_-_12491608 | 0.14 |
ENST00000545735.1
|
MANSC1
|
MANSC domain containing 1 |
| chr2_+_98703595 | 0.13 |
ENST00000435344.1
|
VWA3B
|
von Willebrand factor A domain containing 3B |
| chr3_+_184056614 | 0.13 |
ENST00000453072.1
|
FAM131A
|
family with sequence similarity 131, member A |
| chrX_-_77150911 | 0.12 |
ENST00000373336.3
|
MAGT1
|
magnesium transporter 1 |
| chr6_-_66417107 | 0.12 |
ENST00000370621.3
ENST00000370618.3 ENST00000503581.1 ENST00000393380.2 |
EYS
|
eyes shut homolog (Drosophila) |
| chr2_+_196521903 | 0.12 |
ENST00000541054.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr12_-_95510743 | 0.12 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr9_+_135457530 | 0.11 |
ENST00000263610.2
|
BARHL1
|
BarH-like homeobox 1 |
| chr1_+_161736072 | 0.11 |
ENST00000367942.3
|
ATF6
|
activating transcription factor 6 |
| chr1_-_170043709 | 0.11 |
ENST00000367767.1
ENST00000361580.2 ENST00000538366.1 |
KIFAP3
|
kinesin-associated protein 3 |
| chr2_-_25194476 | 0.10 |
ENST00000534855.1
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr7_+_73868439 | 0.09 |
ENST00000424337.2
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr1_-_232598163 | 0.09 |
ENST00000308942.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chr1_-_160232197 | 0.08 |
ENST00000419626.1
ENST00000610139.1 ENST00000475733.1 ENST00000407642.2 ENST00000368073.3 ENST00000326837.2 |
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr10_+_122610687 | 0.08 |
ENST00000263461.6
|
WDR11
|
WD repeat domain 11 |
| chr4_+_619386 | 0.08 |
ENST00000496514.1
|
PDE6B
|
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chr15_+_67430339 | 0.08 |
ENST00000439724.3
|
SMAD3
|
SMAD family member 3 |
| chr4_+_619347 | 0.07 |
ENST00000255622.6
|
PDE6B
|
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chr5_-_68665296 | 0.06 |
ENST00000512152.1
ENST00000503245.1 ENST00000512561.1 ENST00000380822.4 |
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chr3_+_193853927 | 0.06 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr1_-_160232291 | 0.06 |
ENST00000368074.1
ENST00000447377.1 |
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr3_-_27410847 | 0.05 |
ENST00000429845.2
ENST00000341435.5 ENST00000435750.1 |
NEK10
|
NIMA-related kinase 10 |
| chr6_-_137314371 | 0.05 |
ENST00000432330.1
ENST00000418699.1 |
RP11-55K22.5
|
RP11-55K22.5 |
| chr9_-_124976154 | 0.04 |
ENST00000482062.1
|
LHX6
|
LIM homeobox 6 |
| chr3_+_187461442 | 0.04 |
ENST00000450760.1
|
RP11-211G3.2
|
RP11-211G3.2 |
| chr11_-_119234876 | 0.04 |
ENST00000525735.1
|
USP2
|
ubiquitin specific peptidase 2 |
| chr20_+_31749574 | 0.04 |
ENST00000253362.2
|
BPIFA2
|
BPI fold containing family A, member 2 |
| chr15_-_58358607 | 0.04 |
ENST00000249750.4
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr17_-_60142609 | 0.04 |
ENST00000397786.2
|
MED13
|
mediator complex subunit 13 |
| chr2_-_179914760 | 0.03 |
ENST00000420890.2
ENST00000409284.1 ENST00000443758.1 ENST00000446116.1 |
CCDC141
|
coiled-coil domain containing 141 |
| chr18_+_19749386 | 0.03 |
ENST00000269216.3
|
GATA6
|
GATA binding protein 6 |
| chr8_+_97773202 | 0.03 |
ENST00000519484.1
|
CPQ
|
carboxypeptidase Q |
| chr11_-_110583912 | 0.02 |
ENST00000533353.1
ENST00000527598.1 |
ARHGAP20
|
Rho GTPase activating protein 20 |
| chr17_-_8702667 | 0.02 |
ENST00000329805.4
|
MFSD6L
|
major facilitator superfamily domain containing 6-like |
| chr10_-_95504 | 0.02 |
ENST00000447903.2
|
TUBB8
|
tubulin, beta 8 class VIII |
| chr5_-_142023766 | 0.02 |
ENST00000394496.2
|
FGF1
|
fibroblast growth factor 1 (acidic) |
| chr9_+_35792151 | 0.02 |
ENST00000342694.2
|
NPR2
|
natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B) |
| chr6_+_12717892 | 0.02 |
ENST00000379350.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr8_+_37553261 | 0.01 |
ENST00000331569.4
|
ZNF703
|
zinc finger protein 703 |
| chr1_+_63063152 | 0.01 |
ENST00000371129.3
|
ANGPTL3
|
angiopoietin-like 3 |
| chr9_-_86593238 | 0.01 |
ENST00000351839.3
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K |
| chr14_-_94942548 | 0.01 |
ENST00000298845.7
|
SERPINA9
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 9 |
| chr17_-_41738931 | 0.00 |
ENST00000329168.3
ENST00000549132.1 |
MEOX1
|
mesenchyme homeobox 1 |
| chr8_+_97773457 | 0.00 |
ENST00000521142.1
|
CPQ
|
carboxypeptidase Q |
Network of associatons between targets according to the STRING database.
First level regulatory network of BRCA1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.2 | 1.3 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.2 | 1.0 | GO:1901545 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.1 | 0.9 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.1 | 2.0 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 0.4 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 | 0.6 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.1 | 0.5 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.5 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 0.4 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 0.8 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.2 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.1 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.0 | 0.2 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.7 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) negative regulation of auditory receptor cell differentiation(GO:0045608) regulation of timing of neuron differentiation(GO:0060164) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.5 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 0.5 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.4 | GO:0034380 | cartilage condensation(GO:0001502) high-density lipoprotein particle assembly(GO:0034380) |
| 0.0 | 0.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 0.9 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.5 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.6 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.4 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:1990075 | kinesin II complex(GO:0016939) periciliary membrane compartment(GO:1990075) |
| 0.0 | 2.0 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.1 | 0.7 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 1.3 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.8 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 1.0 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.5 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 3.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.6 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 2.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 2.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.7 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |