Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
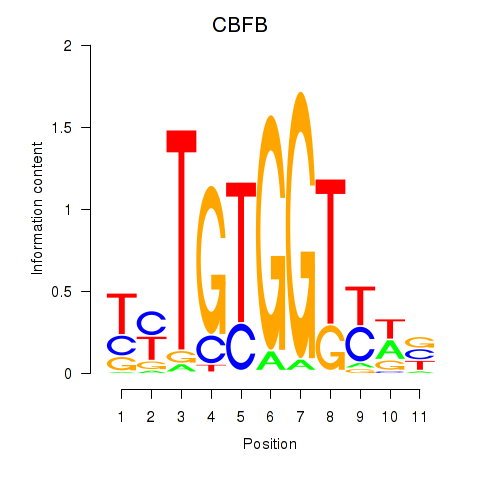
Results for CBFB
Z-value: 0.91
Transcription factors associated with CBFB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CBFB
|
ENSG00000067955.9 | core-binding factor subunit beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CBFB | hg19_v2_chr16_+_67063855_67063873 | 0.94 | 5.3e-10 | Click! |
Activity profile of CBFB motif
Sorted Z-values of CBFB motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_209859510 | 4.58 |
ENST00000367028.2
ENST00000261465.1 |
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr20_+_44637526 | 3.62 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr14_+_85996507 | 3.48 |
ENST00000554746.1
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr14_+_85996471 | 3.21 |
ENST00000330753.4
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr6_-_75912508 | 3.00 |
ENST00000416123.2
|
COL12A1
|
collagen, type XII, alpha 1 |
| chrX_+_115567767 | 2.43 |
ENST00000371900.4
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14 |
| chr3_-_99569821 | 2.34 |
ENST00000487087.1
|
FILIP1L
|
filamin A interacting protein 1-like |
| chrX_+_68048803 | 2.16 |
ENST00000204961.4
|
EFNB1
|
ephrin-B1 |
| chr6_-_113953705 | 2.13 |
ENST00000452675.1
|
RP11-367G18.1
|
RP11-367G18.1 |
| chr5_-_39203093 | 2.11 |
ENST00000515010.1
|
FYB
|
FYN binding protein |
| chr11_-_58343319 | 1.95 |
ENST00000395074.2
|
LPXN
|
leupaxin |
| chr19_-_14048804 | 1.87 |
ENST00000254320.3
ENST00000586075.1 |
PODNL1
|
podocan-like 1 |
| chr15_-_64665911 | 1.87 |
ENST00000606793.1
ENST00000561349.1 ENST00000560278.1 |
CTD-2116N17.1
|
Uncharacterized protein |
| chr14_-_85996332 | 1.64 |
ENST00000380722.1
|
RP11-497E19.1
|
RP11-497E19.1 |
| chr3_+_156799587 | 1.61 |
ENST00000469196.1
|
RP11-6F2.5
|
RP11-6F2.5 |
| chr5_+_35856951 | 1.58 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr15_+_49715449 | 1.57 |
ENST00000560979.1
|
FGF7
|
fibroblast growth factor 7 |
| chr10_-_29811456 | 1.55 |
ENST00000535393.1
|
SVIL
|
supervillin |
| chr13_+_76210448 | 1.53 |
ENST00000377499.5
|
LMO7
|
LIM domain 7 |
| chr6_+_42847649 | 1.53 |
ENST00000424341.2
ENST00000602561.1 |
RPL7L1
|
ribosomal protein L7-like 1 |
| chr15_+_67390920 | 1.44 |
ENST00000559092.1
ENST00000560175.1 |
SMAD3
|
SMAD family member 3 |
| chr6_+_45296391 | 1.37 |
ENST00000371436.6
ENST00000576263.1 |
RUNX2
|
runt-related transcription factor 2 |
| chr1_+_89246647 | 1.35 |
ENST00000544045.1
|
PKN2
|
protein kinase N2 |
| chr11_+_114310102 | 1.33 |
ENST00000265881.5
|
REXO2
|
RNA exonuclease 2 |
| chr12_-_54778471 | 1.32 |
ENST00000550120.1
ENST00000394313.2 ENST00000547210.1 |
ZNF385A
|
zinc finger protein 385A |
| chr12_-_54778444 | 1.30 |
ENST00000551771.1
|
ZNF385A
|
zinc finger protein 385A |
| chr12_+_25205446 | 1.30 |
ENST00000557489.1
ENST00000354454.3 ENST00000536173.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr14_+_72064945 | 1.30 |
ENST00000537413.1
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1 |
| chr12_-_54778244 | 1.29 |
ENST00000549937.1
|
ZNF385A
|
zinc finger protein 385A |
| chr15_+_73976545 | 1.14 |
ENST00000318443.5
ENST00000537340.2 ENST00000318424.5 |
CD276
|
CD276 molecule |
| chr11_+_114310164 | 1.12 |
ENST00000544196.1
ENST00000539754.1 ENST00000539275.1 |
REXO2
|
RNA exonuclease 2 |
| chr1_+_47264711 | 1.11 |
ENST00000371923.4
ENST00000271153.4 ENST00000371919.4 |
CYP4B1
|
cytochrome P450, family 4, subfamily B, polypeptide 1 |
| chr5_+_131409476 | 1.08 |
ENST00000296871.2
|
CSF2
|
colony stimulating factor 2 (granulocyte-macrophage) |
| chr12_+_25205568 | 0.96 |
ENST00000548766.1
ENST00000556887.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr4_+_74347400 | 0.93 |
ENST00000226355.3
|
AFM
|
afamin |
| chr11_-_66104237 | 0.92 |
ENST00000530056.1
|
RIN1
|
Ras and Rab interactor 1 |
| chr11_-_66103932 | 0.92 |
ENST00000311320.4
|
RIN1
|
Ras and Rab interactor 1 |
| chr1_-_179112189 | 0.83 |
ENST00000512653.1
ENST00000344730.3 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr5_-_108745689 | 0.83 |
ENST00000361189.2
|
PJA2
|
praja ring finger 2, E3 ubiquitin protein ligase |
| chr11_-_66103867 | 0.81 |
ENST00000424433.2
|
RIN1
|
Ras and Rab interactor 1 |
| chr2_-_127977654 | 0.81 |
ENST00000409327.1
|
CYP27C1
|
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr14_-_61116168 | 0.79 |
ENST00000247182.6
|
SIX1
|
SIX homeobox 1 |
| chr12_+_25205155 | 0.78 |
ENST00000550945.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr5_+_67576109 | 0.77 |
ENST00000522084.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr11_+_18343800 | 0.72 |
ENST00000453096.2
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa |
| chr12_-_4754339 | 0.71 |
ENST00000228850.1
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chr4_-_156297949 | 0.70 |
ENST00000515654.1
|
MAP9
|
microtubule-associated protein 9 |
| chr9_+_15422702 | 0.69 |
ENST00000380821.3
ENST00000421710.1 |
SNAPC3
|
small nuclear RNA activating complex, polypeptide 3, 50kDa |
| chr2_-_207024233 | 0.69 |
ENST00000423725.1
ENST00000233190.6 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr8_+_28747884 | 0.67 |
ENST00000287701.10
ENST00000444075.1 ENST00000403668.2 ENST00000519662.1 ENST00000558662.1 ENST00000523613.1 ENST00000560599.1 ENST00000397358.3 |
HMBOX1
|
homeobox containing 1 |
| chr2_-_64881018 | 0.65 |
ENST00000313349.3
|
SERTAD2
|
SERTA domain containing 2 |
| chr19_-_14049184 | 0.61 |
ENST00000339560.5
|
PODNL1
|
podocan-like 1 |
| chr16_+_53088885 | 0.61 |
ENST00000566029.1
ENST00000447540.1 |
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr18_-_53089723 | 0.58 |
ENST00000561992.1
ENST00000562512.2 |
TCF4
|
transcription factor 4 |
| chr15_-_33447055 | 0.58 |
ENST00000559047.1
ENST00000561249.1 |
FMN1
|
formin 1 |
| chr5_-_137878887 | 0.55 |
ENST00000507939.1
ENST00000572514.1 ENST00000499810.2 ENST00000360541.5 |
ETF1
|
eukaryotic translation termination factor 1 |
| chr11_+_114310237 | 0.55 |
ENST00000539119.1
|
REXO2
|
RNA exonuclease 2 |
| chr12_-_53594227 | 0.47 |
ENST00000550743.2
|
ITGB7
|
integrin, beta 7 |
| chr21_-_36421626 | 0.46 |
ENST00000300305.3
|
RUNX1
|
runt-related transcription factor 1 |
| chr12_-_4754356 | 0.41 |
ENST00000540967.1
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chr1_-_179112173 | 0.32 |
ENST00000408940.3
ENST00000504405.1 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr12_-_4754318 | 0.31 |
ENST00000536414.1
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chr13_-_103019744 | 0.31 |
ENST00000437115.2
|
FGF14-IT1
|
FGF14 intronic transcript 1 (non-protein coding) |
| chr1_+_39876151 | 0.29 |
ENST00000530275.1
|
KIAA0754
|
KIAA0754 |
| chr1_+_196621156 | 0.28 |
ENST00000359637.2
|
CFH
|
complement factor H |
| chr21_-_36421535 | 0.28 |
ENST00000416754.1
ENST00000437180.1 ENST00000455571.1 |
RUNX1
|
runt-related transcription factor 1 |
| chr2_+_207024306 | 0.28 |
ENST00000236957.5
ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr1_+_248058889 | 0.25 |
ENST00000360358.3
|
OR2W3
|
olfactory receptor, family 2, subfamily W, member 3 |
| chr14_-_24977457 | 0.25 |
ENST00000250378.3
ENST00000206446.4 |
CMA1
|
chymase 1, mast cell |
| chr21_-_36421401 | 0.24 |
ENST00000486278.2
|
RUNX1
|
runt-related transcription factor 1 |
| chr1_-_184723942 | 0.24 |
ENST00000318130.8
|
EDEM3
|
ER degradation enhancer, mannosidase alpha-like 3 |
| chr11_-_18343725 | 0.21 |
ENST00000531848.1
|
HPS5
|
Hermansky-Pudlak syndrome 5 |
| chr2_+_68592305 | 0.20 |
ENST00000234313.7
|
PLEK
|
pleckstrin |
| chr22_+_42665742 | 0.20 |
ENST00000332965.3
ENST00000415205.1 ENST00000446578.1 |
Z83851.3
|
Z83851.3 |
| chr1_+_159770292 | 0.16 |
ENST00000536257.1
ENST00000321935.6 |
FCRL6
|
Fc receptor-like 6 |
| chr18_-_53089538 | 0.16 |
ENST00000566777.1
|
TCF4
|
transcription factor 4 |
| chr15_+_64386261 | 0.16 |
ENST00000560829.1
|
SNX1
|
sorting nexin 1 |
| chrX_-_49041242 | 0.14 |
ENST00000453382.1
ENST00000540849.1 ENST00000536904.1 ENST00000432913.1 |
PRICKLE3
|
prickle homolog 3 (Drosophila) |
| chr3_+_187957646 | 0.13 |
ENST00000457242.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr11_-_1912084 | 0.13 |
ENST00000391480.1
|
C11orf89
|
chromosome 11 open reading frame 89 |
| chr9_-_123342415 | 0.13 |
ENST00000349780.4
ENST00000360190.4 ENST00000360822.3 ENST00000359309.3 |
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2 |
| chr11_-_18343669 | 0.12 |
ENST00000396253.3
ENST00000349215.3 ENST00000438420.2 |
HPS5
|
Hermansky-Pudlak syndrome 5 |
| chr13_-_28674693 | 0.10 |
ENST00000537084.1
ENST00000241453.7 ENST00000380982.4 |
FLT3
|
fms-related tyrosine kinase 3 |
| chr8_-_28747717 | 0.07 |
ENST00000416984.2
|
INTS9
|
integrator complex subunit 9 |
| chr12_-_14849470 | 0.06 |
ENST00000261170.3
|
GUCY2C
|
guanylate cyclase 2C (heat stable enterotoxin receptor) |
| chr20_+_54987168 | 0.04 |
ENST00000360314.3
|
CASS4
|
Cas scaffolding protein family member 4 |
| chr8_-_28747424 | 0.03 |
ENST00000523436.1
ENST00000397363.4 ENST00000521777.1 ENST00000520184.1 ENST00000521022.1 |
INTS9
|
integrator complex subunit 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CBFB
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 6.7 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.6 | 5.1 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.6 | 3.9 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.4 | 1.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.3 | 2.7 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.2 | 1.4 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.2 | 0.8 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.2 | 1.9 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.2 | 4.6 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.1 | 1.6 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 1.1 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.1 | 1.5 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 1.4 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.5 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.1 | 1.1 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.1 | 0.3 | GO:0050720 | interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.1 | 0.6 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 1.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 2.9 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.7 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.1 | 3.0 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 2.1 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 3.0 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 2.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.6 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.8 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.2 | GO:0097466 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) glycoprotein ERAD pathway(GO:0097466) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.0 | 2.2 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 0.2 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.0 | 2.5 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.8 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 0.7 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.7 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 3.1 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 2.4 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 1.1 | GO:0010863 | positive regulation of phospholipase C activity(GO:0010863) |
| 0.0 | 1.3 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 0.9 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.1 | GO:0035726 | myeloid progenitor cell differentiation(GO:0002318) common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.1 | 1.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.7 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.1 | 1.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.5 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.1 | 1.5 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.8 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 0.7 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.1 | 0.3 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 0.7 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 3.2 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 3.1 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 1.9 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 3.0 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 2.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 1.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.4 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 6.0 | GO:0000790 | nuclear chromatin(GO:0000790) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.4 | 1.6 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.3 | 6.7 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.3 | 3.0 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.2 | 0.9 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.2 | 1.4 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.2 | 0.8 | GO:0005502 | 11-cis retinal binding(GO:0005502) all-trans retinal binding(GO:0005503) |
| 0.1 | 2.4 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 0.8 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 0.6 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 1.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 3.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 3.0 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 1.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.7 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.7 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 1.4 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.8 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 2.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 3.9 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.7 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 3.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.8 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 1.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 1.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.4 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 2.5 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 2.9 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 2.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 2.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 2.1 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 1.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 2.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 4.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 2.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 2.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.5 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 2.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.1 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 2.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.7 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.1 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.7 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 1.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |