Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
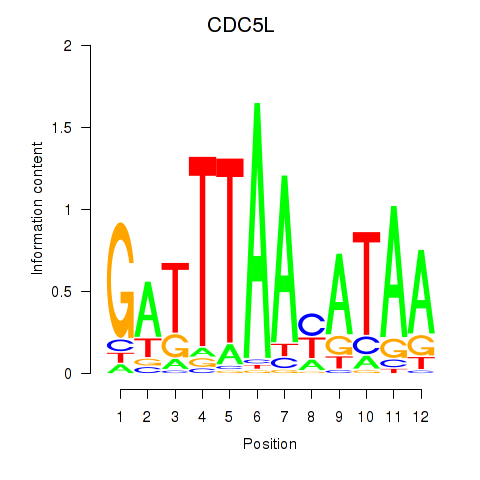
Results for CDC5L
Z-value: 0.52
Transcription factors associated with CDC5L
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CDC5L
|
ENSG00000096401.7 | cell division cycle 5 like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CDC5L | hg19_v2_chr6_+_44355257_44355315 | -0.44 | 5.3e-02 | Click! |
Activity profile of CDC5L motif
Sorted Z-values of CDC5L motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_155484155 | 1.66 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr10_-_69597810 | 1.54 |
ENST00000483798.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr4_+_155484103 | 1.49 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr10_-_69597915 | 1.47 |
ENST00000225171.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr10_-_69597828 | 1.22 |
ENST00000339758.7
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr8_-_124749609 | 0.99 |
ENST00000262219.6
ENST00000419625.1 |
ANXA13
|
annexin A13 |
| chr10_+_91152303 | 0.67 |
ENST00000371804.3
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1 |
| chr3_+_151531859 | 0.54 |
ENST00000488869.1
|
AADAC
|
arylacetamide deacetylase |
| chr17_-_64225508 | 0.52 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr4_-_110723194 | 0.50 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr4_-_110723134 | 0.49 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr4_+_80584903 | 0.48 |
ENST00000506460.1
|
RP11-452C8.1
|
RP11-452C8.1 |
| chr3_-_120365866 | 0.48 |
ENST00000475447.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr3_+_151531810 | 0.46 |
ENST00000232892.7
|
AADAC
|
arylacetamide deacetylase |
| chrX_-_15402498 | 0.45 |
ENST00000297904.3
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D) |
| chr2_+_228678550 | 0.43 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr2_+_232573208 | 0.42 |
ENST00000409115.3
|
PTMA
|
prothymosin, alpha |
| chr8_+_52730143 | 0.42 |
ENST00000415643.1
|
AC090186.1
|
Uncharacterized protein |
| chr21_+_33784670 | 0.41 |
ENST00000300255.2
|
EVA1C
|
eva-1 homolog C (C. elegans) |
| chr1_+_196621002 | 0.38 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr15_-_42076229 | 0.35 |
ENST00000597767.1
|
AC073657.1
|
Uncharacterized protein |
| chr10_+_96698406 | 0.35 |
ENST00000260682.6
|
CYP2C9
|
cytochrome P450, family 2, subfamily C, polypeptide 9 |
| chr1_-_150738261 | 0.35 |
ENST00000448301.2
ENST00000368985.3 |
CTSS
|
cathepsin S |
| chr2_+_232573222 | 0.34 |
ENST00000341369.7
ENST00000409683.1 |
PTMA
|
prothymosin, alpha |
| chr8_+_76452097 | 0.32 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr9_+_40028620 | 0.31 |
ENST00000426179.1
|
AL353791.1
|
AL353791.1 |
| chr12_+_20963647 | 0.31 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr14_-_54425475 | 0.31 |
ENST00000559642.1
|
BMP4
|
bone morphogenetic protein 4 |
| chr16_-_71610985 | 0.30 |
ENST00000355962.4
|
TAT
|
tyrosine aminotransferase |
| chr3_-_119396193 | 0.29 |
ENST00000484810.1
ENST00000497116.1 ENST00000261070.2 |
COX17
|
COX17 cytochrome c oxidase copper chaperone |
| chr14_-_92247032 | 0.28 |
ENST00000556661.1
ENST00000553676.1 ENST00000554560.1 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr2_+_200472779 | 0.28 |
ENST00000427045.1
ENST00000419243.1 |
AC093590.1
|
AC093590.1 |
| chr17_-_15469590 | 0.28 |
ENST00000312127.2
|
CDRT1
|
CMT duplicated region transcript 1; Uncharacterized protein |
| chr1_-_89591749 | 0.28 |
ENST00000370466.3
|
GBP2
|
guanylate binding protein 2, interferon-inducible |
| chr13_+_78315295 | 0.28 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr4_-_23735183 | 0.28 |
ENST00000507666.1
|
RP11-380P13.2
|
RP11-380P13.2 |
| chr3_+_171561127 | 0.27 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr1_+_70820451 | 0.27 |
ENST00000361764.4
ENST00000359875.5 ENST00000370940.5 ENST00000531950.1 ENST00000432224.1 |
HHLA3
|
HERV-H LTR-associating 3 |
| chr9_+_35673853 | 0.27 |
ENST00000378357.4
|
CA9
|
carbonic anhydrase IX |
| chr1_+_95616933 | 0.26 |
ENST00000604203.1
|
RP11-57H12.6
|
TMEM56-RWDD3 readthrough |
| chr11_+_57308979 | 0.26 |
ENST00000457912.1
|
SMTNL1
|
smoothelin-like 1 |
| chr11_+_73661364 | 0.26 |
ENST00000339764.1
|
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr7_+_5465382 | 0.25 |
ENST00000609130.1
|
RP11-1275H24.2
|
RP11-1275H24.2 |
| chr6_+_32812568 | 0.25 |
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr12_+_20963632 | 0.25 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr6_+_27791862 | 0.25 |
ENST00000355057.1
|
HIST1H4J
|
histone cluster 1, H4j |
| chr9_-_21368075 | 0.25 |
ENST00000449498.1
|
IFNA13
|
interferon, alpha 13 |
| chr15_-_55562582 | 0.25 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr19_+_782755 | 0.24 |
ENST00000606242.1
ENST00000586061.1 |
AC006273.5
|
AC006273.5 |
| chr14_-_38036271 | 0.24 |
ENST00000556024.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr9_-_21482312 | 0.24 |
ENST00000448696.3
|
IFNE
|
interferon, epsilon |
| chr1_-_65468148 | 0.24 |
ENST00000415842.1
|
RP11-182I10.3
|
RP11-182I10.3 |
| chr2_-_190445499 | 0.24 |
ENST00000261024.2
|
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr22_-_29107919 | 0.24 |
ENST00000434810.1
ENST00000456369.1 |
CHEK2
|
checkpoint kinase 2 |
| chr10_+_124739964 | 0.23 |
ENST00000406217.2
|
PSTK
|
phosphoseryl-tRNA kinase |
| chr17_+_18647326 | 0.23 |
ENST00000395667.1
ENST00000395665.4 ENST00000308799.4 ENST00000301938.4 |
FBXW10
|
F-box and WD repeat domain containing 10 |
| chr2_+_132479948 | 0.23 |
ENST00000355171.4
|
C2orf27A
|
chromosome 2 open reading frame 27A |
| chrX_-_11129229 | 0.23 |
ENST00000608176.1
ENST00000433747.2 ENST00000608576.1 ENST00000608916.1 |
RP11-120D5.1
|
RP11-120D5.1 |
| chr1_+_198608146 | 0.23 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr8_+_35649365 | 0.23 |
ENST00000437887.1
|
AC012215.1
|
Uncharacterized protein |
| chr10_+_5135981 | 0.23 |
ENST00000380554.3
|
AKR1C3
|
aldo-keto reductase family 1, member C3 |
| chr8_-_110988070 | 0.23 |
ENST00000524391.1
|
KCNV1
|
potassium channel, subfamily V, member 1 |
| chr6_+_26156551 | 0.22 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr2_-_677369 | 0.22 |
ENST00000281017.3
|
TMEM18
|
transmembrane protein 18 |
| chr5_+_43033818 | 0.22 |
ENST00000607830.1
|
CTD-2035E11.4
|
CTD-2035E11.4 |
| chr12_+_100897130 | 0.22 |
ENST00000551379.1
ENST00000188403.7 ENST00000551184.1 |
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr5_-_179499108 | 0.22 |
ENST00000521389.1
|
RNF130
|
ring finger protein 130 |
| chr4_+_74606223 | 0.21 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr6_+_136172820 | 0.21 |
ENST00000308191.6
|
PDE7B
|
phosphodiesterase 7B |
| chr12_+_58003935 | 0.21 |
ENST00000333972.7
|
ARHGEF25
|
Rho guanine nucleotide exchange factor (GEF) 25 |
| chr11_-_31531121 | 0.21 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr1_+_66458072 | 0.21 |
ENST00000423207.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr4_-_130692631 | 0.21 |
ENST00000500092.2
ENST00000509105.1 |
RP11-519M16.1
|
RP11-519M16.1 |
| chr1_+_244998602 | 0.20 |
ENST00000411948.2
|
COX20
|
COX20 cytochrome C oxidase assembly factor |
| chr1_+_10509971 | 0.20 |
ENST00000320498.4
|
CORT
|
cortistatin |
| chr16_-_4401258 | 0.20 |
ENST00000577031.1
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr4_+_164265035 | 0.19 |
ENST00000338566.3
|
NPY5R
|
neuropeptide Y receptor Y5 |
| chr12_-_91546926 | 0.19 |
ENST00000550758.1
|
DCN
|
decorin |
| chr19_-_9006766 | 0.19 |
ENST00000599436.1
|
MUC16
|
mucin 16, cell surface associated |
| chr18_+_61442629 | 0.19 |
ENST00000398019.2
ENST00000540675.1 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr19_+_852291 | 0.19 |
ENST00000263621.1
|
ELANE
|
elastase, neutrophil expressed |
| chr14_+_76452090 | 0.18 |
ENST00000314067.6
ENST00000238628.6 ENST00000556742.1 |
IFT43
|
intraflagellar transport 43 homolog (Chlamydomonas) |
| chr5_+_139505520 | 0.18 |
ENST00000333305.3
|
IGIP
|
IgA-inducing protein |
| chr18_+_3466248 | 0.18 |
ENST00000581029.1
ENST00000581442.1 ENST00000579007.1 |
RP11-838N2.4
|
RP11-838N2.4 |
| chr14_+_104029362 | 0.18 |
ENST00000495778.1
|
APOPT1
|
apoptogenic 1, mitochondrial |
| chr8_-_101719159 | 0.18 |
ENST00000520868.1
ENST00000522658.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr12_+_4714145 | 0.18 |
ENST00000545342.1
|
DYRK4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr2_+_74685413 | 0.17 |
ENST00000233615.2
|
WBP1
|
WW domain binding protein 1 |
| chr3_-_149051444 | 0.17 |
ENST00000296059.2
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chrX_-_23926004 | 0.17 |
ENST00000379226.4
ENST00000379220.3 |
APOO
|
apolipoprotein O |
| chr16_+_80574854 | 0.17 |
ENST00000305904.6
ENST00000568035.1 |
DYNLRB2
|
dynein, light chain, roadblock-type 2 |
| chr2_+_85132749 | 0.17 |
ENST00000233143.4
|
TMSB10
|
thymosin beta 10 |
| chr15_+_35270552 | 0.17 |
ENST00000391457.2
|
AC114546.1
|
HCG37415; PRO1914; Uncharacterized protein |
| chr17_+_45908974 | 0.17 |
ENST00000269025.4
|
LRRC46
|
leucine rich repeat containing 46 |
| chrX_+_100646190 | 0.16 |
ENST00000471855.1
|
RPL36A
|
ribosomal protein L36a |
| chr1_+_196621156 | 0.16 |
ENST00000359637.2
|
CFH
|
complement factor H |
| chr7_-_16921601 | 0.16 |
ENST00000402239.3
ENST00000310398.2 ENST00000414935.1 |
AGR3
|
anterior gradient 3 |
| chr1_-_89641680 | 0.16 |
ENST00000294671.2
|
GBP7
|
guanylate binding protein 7 |
| chr9_-_4666421 | 0.16 |
ENST00000381895.5
|
SPATA6L
|
spermatogenesis associated 6-like |
| chr19_-_11545920 | 0.15 |
ENST00000356392.4
ENST00000591179.1 |
CCDC151
|
coiled-coil domain containing 151 |
| chr12_+_20968608 | 0.15 |
ENST00000381541.3
ENST00000540229.1 ENST00000553473.1 ENST00000554957.1 |
LST3
SLCO1B3
SLCO1B7
|
Putative solute carrier organic anion transporter family member 1B7; Uncharacterized protein solute carrier organic anion transporter family, member 1B3 solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr2_+_54350316 | 0.15 |
ENST00000606865.1
|
ACYP2
|
acylphosphatase 2, muscle type |
| chr2_-_190927447 | 0.15 |
ENST00000260950.4
|
MSTN
|
myostatin |
| chr1_+_241695670 | 0.15 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr18_+_616711 | 0.15 |
ENST00000579494.1
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr8_-_133637624 | 0.15 |
ENST00000522789.1
|
LRRC6
|
leucine rich repeat containing 6 |
| chr17_-_8263538 | 0.15 |
ENST00000535173.1
|
AC135178.1
|
HCG1985372; Uncharacterized protein; cDNA FLJ37541 fis, clone BRCAN2026340 |
| chr1_-_154178803 | 0.15 |
ENST00000368525.3
|
C1orf189
|
chromosome 1 open reading frame 189 |
| chr12_-_49463620 | 0.15 |
ENST00000550675.1
|
RHEBL1
|
Ras homolog enriched in brain like 1 |
| chr11_+_64794875 | 0.15 |
ENST00000377244.3
ENST00000534637.1 ENST00000524831.1 |
SNX15
|
sorting nexin 15 |
| chr11_-_32816156 | 0.15 |
ENST00000531481.1
ENST00000335185.5 |
CCDC73
|
coiled-coil domain containing 73 |
| chr1_+_22351977 | 0.15 |
ENST00000420503.1
ENST00000416769.1 ENST00000404210.2 |
LINC00339
|
long intergenic non-protein coding RNA 339 |
| chr10_+_135207598 | 0.15 |
ENST00000477902.2
|
MTG1
|
mitochondrial ribosome-associated GTPase 1 |
| chr12_-_21928515 | 0.14 |
ENST00000537950.1
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr20_+_16710606 | 0.14 |
ENST00000377943.5
ENST00000246071.6 |
SNRPB2
|
small nuclear ribonucleoprotein polypeptide B |
| chr16_+_33204156 | 0.14 |
ENST00000398667.4
|
TP53TG3C
|
TP53 target 3C |
| chr5_-_95018660 | 0.14 |
ENST00000395899.3
ENST00000274432.8 |
SPATA9
|
spermatogenesis associated 9 |
| chr1_+_229440129 | 0.14 |
ENST00000366688.3
|
SPHAR
|
S-phase response (cyclin related) |
| chr6_+_46661575 | 0.14 |
ENST00000450697.1
|
TDRD6
|
tudor domain containing 6 |
| chr2_-_74570520 | 0.14 |
ENST00000394019.2
ENST00000346834.4 ENST00000359484.4 ENST00000423644.1 ENST00000377634.4 ENST00000436454.1 |
SLC4A5
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 5 |
| chr11_-_62609281 | 0.14 |
ENST00000525239.1
ENST00000538098.2 |
WDR74
|
WD repeat domain 74 |
| chr4_+_75023816 | 0.14 |
ENST00000395759.2
ENST00000331145.6 ENST00000359107.5 ENST00000325278.6 |
MTHFD2L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like |
| chr22_-_39268308 | 0.14 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr19_-_33360647 | 0.14 |
ENST00000590341.1
ENST00000587772.1 ENST00000023064.4 |
SLC7A9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr5_+_140213815 | 0.14 |
ENST00000525929.1
ENST00000378125.3 |
PCDHA7
|
protocadherin alpha 7 |
| chr8_-_102216925 | 0.14 |
ENST00000517844.1
|
ZNF706
|
zinc finger protein 706 |
| chr1_+_43613612 | 0.14 |
ENST00000335282.4
|
FAM183A
|
family with sequence similarity 183, member A |
| chr9_+_21440440 | 0.14 |
ENST00000276927.1
|
IFNA1
|
interferon, alpha 1 |
| chr21_+_35107346 | 0.14 |
ENST00000456489.1
|
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr7_+_66800928 | 0.13 |
ENST00000430244.1
|
RP11-166O4.5
|
RP11-166O4.5 |
| chr15_+_75970672 | 0.13 |
ENST00000435356.1
|
AC105020.1
|
Uncharacterized protein; cDNA FLJ12988 fis, clone NT2RP3000080 |
| chr7_-_102985288 | 0.13 |
ENST00000379263.3
|
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr16_+_22019404 | 0.13 |
ENST00000542527.2
ENST00000569656.1 ENST00000562695.1 |
C16orf52
|
chromosome 16 open reading frame 52 |
| chr10_+_124739911 | 0.13 |
ENST00000405485.1
|
PSTK
|
phosphoseryl-tRNA kinase |
| chr20_+_52824367 | 0.13 |
ENST00000371419.2
|
PFDN4
|
prefoldin subunit 4 |
| chr1_-_84972248 | 0.13 |
ENST00000370645.4
ENST00000370641.3 |
GNG5
|
guanine nucleotide binding protein (G protein), gamma 5 |
| chr2_-_27632390 | 0.13 |
ENST00000350803.4
ENST00000344034.4 |
PPM1G
|
protein phosphatase, Mg2+/Mn2+ dependent, 1G |
| chr1_+_144339738 | 0.13 |
ENST00000538264.1
|
AL592284.1
|
Protein LOC642441 |
| chr13_+_78315466 | 0.13 |
ENST00000314070.5
ENST00000462234.1 |
SLAIN1
|
SLAIN motif family, member 1 |
| chr4_+_118955500 | 0.13 |
ENST00000296499.5
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr12_+_16500599 | 0.13 |
ENST00000535309.1
ENST00000540056.1 ENST00000396209.1 ENST00000540126.1 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr4_+_119200215 | 0.13 |
ENST00000602573.1
|
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr3_-_155394099 | 0.13 |
ENST00000414191.1
|
PLCH1
|
phospholipase C, eta 1 |
| chr1_-_158173659 | 0.13 |
ENST00000415019.1
|
RP11-404O13.5
|
RP11-404O13.5 |
| chr17_-_73775839 | 0.13 |
ENST00000592643.1
ENST00000591890.1 ENST00000587171.1 ENST00000254810.4 ENST00000589599.1 |
H3F3B
|
H3 histone, family 3B (H3.3B) |
| chr12_-_10542617 | 0.12 |
ENST00000240618.6
|
KLRK1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr6_+_52535878 | 0.12 |
ENST00000211314.4
|
TMEM14A
|
transmembrane protein 14A |
| chr10_+_35484053 | 0.12 |
ENST00000487763.1
ENST00000473940.1 ENST00000488328.1 ENST00000356917.5 |
CREM
|
cAMP responsive element modulator |
| chr19_-_49843539 | 0.12 |
ENST00000602554.1
ENST00000358234.4 |
CTC-301O7.4
|
CTC-301O7.4 |
| chr14_+_39703112 | 0.12 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr3_+_73110810 | 0.12 |
ENST00000533473.1
|
EBLN2
|
endogenous Bornavirus-like nucleoprotein 2 |
| chr18_+_68002675 | 0.12 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr19_-_4540486 | 0.12 |
ENST00000306390.6
|
LRG1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr6_+_27107053 | 0.12 |
ENST00000354348.2
|
HIST1H4I
|
histone cluster 1, H4i |
| chr16_-_71918033 | 0.12 |
ENST00000425432.1
ENST00000313565.6 ENST00000568666.1 ENST00000562797.1 ENST00000564134.1 |
ZNF821
|
zinc finger protein 821 |
| chr4_+_130017268 | 0.12 |
ENST00000425929.1
ENST00000508673.1 ENST00000508622.1 |
C4orf33
|
chromosome 4 open reading frame 33 |
| chr18_+_616672 | 0.12 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr5_-_179499086 | 0.12 |
ENST00000261947.4
|
RNF130
|
ring finger protein 130 |
| chr5_-_127418573 | 0.12 |
ENST00000508353.1
ENST00000508878.1 ENST00000501652.1 ENST00000514409.1 |
CTC-228N24.3
|
CTC-228N24.3 |
| chr12_-_122658746 | 0.12 |
ENST00000377035.1
|
IL31
|
interleukin 31 |
| chr19_+_21579908 | 0.12 |
ENST00000596302.1
ENST00000392288.2 ENST00000594390.1 ENST00000355504.4 |
ZNF493
|
zinc finger protein 493 |
| chr9_-_28670283 | 0.12 |
ENST00000379992.2
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr1_+_179335101 | 0.12 |
ENST00000508285.1
ENST00000511889.1 |
AXDND1
|
axonemal dynein light chain domain containing 1 |
| chr1_+_9599540 | 0.11 |
ENST00000302692.6
|
SLC25A33
|
solute carrier family 25 (pyrimidine nucleotide carrier), member 33 |
| chr9_+_136215044 | 0.11 |
ENST00000323345.6
|
RPL7A
|
ribosomal protein L7a |
| chr13_+_78315348 | 0.11 |
ENST00000441784.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr19_+_35939154 | 0.11 |
ENST00000599180.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr15_-_55611306 | 0.11 |
ENST00000563262.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr12_-_21487829 | 0.11 |
ENST00000445053.1
ENST00000452078.1 ENST00000458504.1 ENST00000422327.1 ENST00000421294.1 |
SLCO1A2
|
solute carrier organic anion transporter family, member 1A2 |
| chr6_-_26250835 | 0.11 |
ENST00000446824.2
|
HIST1H3F
|
histone cluster 1, H3f |
| chr14_+_29241910 | 0.11 |
ENST00000399387.4
ENST00000552957.1 ENST00000548213.1 |
C14orf23
|
chromosome 14 open reading frame 23 |
| chr3_-_57326704 | 0.11 |
ENST00000487349.1
ENST00000389601.3 |
ASB14
|
ankyrin repeat and SOCS box containing 14 |
| chr6_+_123038689 | 0.11 |
ENST00000354275.2
ENST00000368446.1 |
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chrM_+_12331 | 0.11 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr5_+_53686658 | 0.11 |
ENST00000512618.1
|
LINC01033
|
long intergenic non-protein coding RNA 1033 |
| chr21_+_17792672 | 0.11 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr6_+_161123270 | 0.11 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
| chr10_+_135207623 | 0.11 |
ENST00000317502.6
ENST00000432508.3 |
MTG1
|
mitochondrial ribosome-associated GTPase 1 |
| chr1_+_156308403 | 0.11 |
ENST00000481479.1
ENST00000368252.1 ENST00000466306.1 ENST00000368251.1 |
TSACC
|
TSSK6 activating co-chaperone |
| chr11_+_114168773 | 0.11 |
ENST00000542647.1
ENST00000545255.1 |
NNMT
|
nicotinamide N-methyltransferase |
| chr1_+_41204506 | 0.11 |
ENST00000525290.1
ENST00000530965.1 ENST00000416859.2 ENST00000308733.5 |
NFYC
|
nuclear transcription factor Y, gamma |
| chr7_+_29519486 | 0.11 |
ENST00000409041.4
|
CHN2
|
chimerin 2 |
| chr8_-_57906362 | 0.11 |
ENST00000262644.4
|
IMPAD1
|
inositol monophosphatase domain containing 1 |
| chr16_-_46782221 | 0.11 |
ENST00000394809.4
|
MYLK3
|
myosin light chain kinase 3 |
| chr16_-_1843720 | 0.11 |
ENST00000415638.3
ENST00000215539.3 |
IGFALS
|
insulin-like growth factor binding protein, acid labile subunit |
| chr2_+_207804278 | 0.10 |
ENST00000272852.3
|
CPO
|
carboxypeptidase O |
| chrX_-_15683147 | 0.10 |
ENST00000380342.3
|
TMEM27
|
transmembrane protein 27 |
| chr4_-_76957214 | 0.10 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chr19_-_39466396 | 0.10 |
ENST00000292852.4
|
FBXO17
|
F-box protein 17 |
| chr9_-_70866572 | 0.10 |
ENST00000417734.1
|
AL353608.1
|
HDCMB45P; Uncharacterized protein |
| chr2_-_114205429 | 0.10 |
ENST00000440617.1
|
AC016745.1
|
HDCMB45P; Uncharacterized protein |
| chr9_+_125512019 | 0.10 |
ENST00000373684.1
ENST00000304720.2 |
OR1L6
|
olfactory receptor, family 1, subfamily L, member 6 |
| chr1_+_241695424 | 0.10 |
ENST00000366558.3
ENST00000366559.4 |
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr8_+_66933779 | 0.10 |
ENST00000276570.5
|
DNAJC5B
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr11_-_84028339 | 0.10 |
ENST00000398301.2
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr13_+_32313658 | 0.10 |
ENST00000380314.1
ENST00000298386.2 |
RXFP2
|
relaxin/insulin-like family peptide receptor 2 |
| chr5_-_127418755 | 0.10 |
ENST00000501702.2
ENST00000501173.2 ENST00000514573.1 ENST00000499346.2 ENST00000606251.1 |
CTC-228N24.3
|
CTC-228N24.3 |
| chr3_-_150421728 | 0.10 |
ENST00000295910.6
ENST00000491361.1 |
FAM194A
|
family with sequence similarity 194, member A |
| chr4_+_187813673 | 0.10 |
ENST00000606881.1
|
RP11-11N5.3
|
RP11-11N5.3 |
| chr4_-_141348999 | 0.10 |
ENST00000325617.5
|
CLGN
|
calmegin |
Network of associatons between targets according to the STRING database.
First level regulatory network of CDC5L
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.2 | 0.7 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.1 | 0.4 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.1 | 0.3 | GO:1903522 | regulation of blood circulation(GO:1903522) |
| 0.1 | 1.0 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 | 0.8 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.2 | GO:1903988 | spleen trabecula formation(GO:0060345) iron cation export(GO:1903414) ferrous iron export(GO:1903988) |
| 0.1 | 0.2 | GO:0072428 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.2 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.1 | 0.3 | GO:2000006 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.1 | 0.4 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.1 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.1 | 0.2 | GO:2001248 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.1 | 0.4 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.1 | 0.3 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.0 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.6 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.0 | 0.3 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.1 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.3 | GO:0045415 | negative regulation of interleukin-8 biosynthetic process(GO:0045415) |
| 0.0 | 0.1 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.2 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.2 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.0 | 0.2 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.2 | GO:1902725 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.1 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.1 | GO:0002879 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.0 | 0.2 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.3 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.2 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.0 | 0.1 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.2 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.4 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.5 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) sarcomerogenesis(GO:0048769) |
| 0.0 | 0.1 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.3 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:0031394 | myoblast migration involved in skeletal muscle regeneration(GO:0014839) positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.9 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.1 | GO:0036023 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.0 | 0.1 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.1 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.1 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.2 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.0 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.2 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.1 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.1 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.2 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.0 | GO:0002818 | intracellular defense response(GO:0002818) |
| 0.0 | 0.1 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.2 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.2 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.2 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.1 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.1 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.2 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.0 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 | 0.1 | GO:2000111 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.0 | 0.0 | GO:0032072 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.3 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.2 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.5 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.4 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.3 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.0 | 0.1 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.0 | GO:0032127 | dense core granule membrane(GO:0032127) microvesicle(GO:1990742) |
| 0.0 | 0.1 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.2 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.1 | 0.5 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.3 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.2 | GO:0097689 | iron channel activity(GO:0097689) |
| 0.1 | 0.2 | GO:0047017 | geranylgeranyl reductase activity(GO:0045550) prostaglandin-F synthase activity(GO:0047017) |
| 0.1 | 0.3 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.1 | 0.2 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.1 | 0.2 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.1 | 0.2 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 0.4 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.2 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.1 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.1 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.0 | 0.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.9 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 1.1 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.1 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.0 | 0.2 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.0 | 0.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.9 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.1 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.1 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.0 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.1 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 1.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 1.9 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.0 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.0 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0052836 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 1.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.8 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |