Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
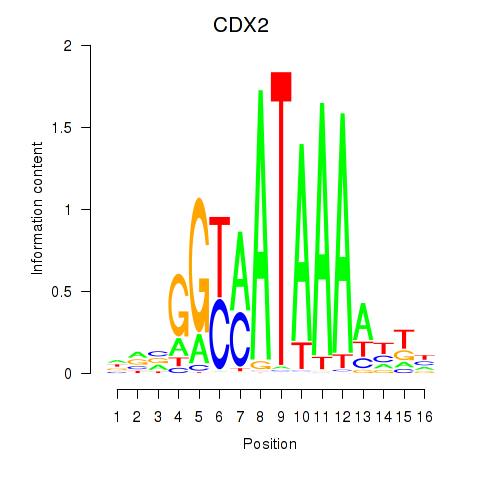
Results for CDX2
Z-value: 0.91
Transcription factors associated with CDX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CDX2
|
ENSG00000165556.9 | caudal type homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CDX2 | hg19_v2_chr13_-_28545276_28545276 | -0.83 | 5.5e-06 | Click! |
Activity profile of CDX2 motif
Sorted Z-values of CDX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_53636125 | 3.94 |
ENST00000601493.1
ENST00000599261.1 ENST00000597503.1 ENST00000500065.4 ENST00000243643.4 ENST00000594011.1 ENST00000455735.2 ENST00000595193.1 ENST00000448501.1 ENST00000421033.1 ENST00000440291.1 ENST00000595813.1 ENST00000600574.1 ENST00000596051.1 ENST00000601110.1 |
ZNF415
|
zinc finger protein 415 |
| chr3_-_172241250 | 3.64 |
ENST00000420541.2
ENST00000241261.2 |
TNFSF10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr4_-_15939963 | 3.51 |
ENST00000259988.2
|
FGFBP1
|
fibroblast growth factor binding protein 1 |
| chr10_-_5660118 | 3.48 |
ENST00000427341.1
|
RP11-336A10.4
|
RP11-336A10.4 |
| chr2_+_158114051 | 3.32 |
ENST00000259056.4
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr1_+_158979792 | 3.30 |
ENST00000359709.3
ENST00000430894.2 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr1_+_158979686 | 3.16 |
ENST00000368132.3
ENST00000295809.7 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr8_+_143761874 | 2.92 |
ENST00000301258.4
ENST00000513264.1 |
PSCA
|
prostate stem cell antigen |
| chrX_+_105936982 | 2.90 |
ENST00000418562.1
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr3_-_18480260 | 2.69 |
ENST00000454909.2
|
SATB1
|
SATB homeobox 1 |
| chr7_-_41742697 | 2.66 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chrX_+_105937068 | 2.43 |
ENST00000324342.3
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr11_+_12302492 | 2.40 |
ENST00000533534.1
|
MICALCL
|
MICAL C-terminal like |
| chr9_-_73029540 | 2.12 |
ENST00000377126.2
|
KLF9
|
Kruppel-like factor 9 |
| chr7_-_143105941 | 2.08 |
ENST00000275815.3
|
EPHA1
|
EPH receptor A1 |
| chr3_-_196911002 | 2.05 |
ENST00000452595.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr11_+_71900703 | 2.01 |
ENST00000393681.2
|
FOLR1
|
folate receptor 1 (adult) |
| chr5_-_39270725 | 1.92 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr17_-_80797886 | 1.90 |
ENST00000572562.1
|
ZNF750
|
zinc finger protein 750 |
| chr11_+_71900572 | 1.86 |
ENST00000312293.4
|
FOLR1
|
folate receptor 1 (adult) |
| chr18_+_61445205 | 1.86 |
ENST00000431370.1
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr1_+_40942887 | 1.76 |
ENST00000372706.1
|
ZFP69
|
ZFP69 zinc finger protein |
| chr12_-_52828147 | 1.66 |
ENST00000252245.5
|
KRT75
|
keratin 75 |
| chrM_+_10053 | 1.65 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr10_+_24755416 | 1.65 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr3_-_69591727 | 1.61 |
ENST00000459638.1
|
FRMD4B
|
FERM domain containing 4B |
| chr4_+_71384300 | 1.55 |
ENST00000504451.1
|
AMTN
|
amelotin |
| chr19_+_21106028 | 1.52 |
ENST00000597314.1
ENST00000601924.1 |
ZNF85
|
zinc finger protein 85 |
| chr1_+_24645865 | 1.50 |
ENST00000342072.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr3_-_18480173 | 1.49 |
ENST00000414509.1
|
SATB1
|
SATB homeobox 1 |
| chr1_+_24645807 | 1.45 |
ENST00000361548.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr4_+_71384257 | 1.41 |
ENST00000339336.4
|
AMTN
|
amelotin |
| chr2_+_210444142 | 1.40 |
ENST00000360351.4
ENST00000361559.4 |
MAP2
|
microtubule-associated protein 2 |
| chrM_+_10464 | 1.36 |
ENST00000361335.1
|
MT-ND4L
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr19_+_21106081 | 1.35 |
ENST00000300540.3
ENST00000595854.1 ENST00000601284.1 ENST00000328178.8 ENST00000599885.1 ENST00000596476.1 ENST00000345030.6 |
ZNF85
|
zinc finger protein 85 |
| chr7_+_5468362 | 1.28 |
ENST00000608012.1
|
RP11-1275H24.3
|
RP11-1275H24.3 |
| chrX_-_15619076 | 1.26 |
ENST00000252519.3
|
ACE2
|
angiotensin I converting enzyme 2 |
| chrX_-_30993201 | 1.24 |
ENST00000288422.2
ENST00000378932.2 |
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr12_+_65996599 | 1.24 |
ENST00000539116.1
ENST00000541391.1 |
RP11-221N13.3
|
RP11-221N13.3 |
| chr2_+_210443993 | 1.23 |
ENST00000392193.1
|
MAP2
|
microtubule-associated protein 2 |
| chr20_-_36152914 | 1.22 |
ENST00000397131.1
|
BLCAP
|
bladder cancer associated protein |
| chr7_+_134464376 | 1.12 |
ENST00000454108.1
ENST00000361675.2 |
CALD1
|
caldesmon 1 |
| chr19_-_43032532 | 1.10 |
ENST00000403461.1
ENST00000352591.5 ENST00000358394.3 ENST00000403444.3 ENST00000308072.4 ENST00000599389.1 ENST00000351134.3 ENST00000161559.6 |
CEACAM1
|
carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) |
| chr21_-_27462351 | 1.07 |
ENST00000448850.1
|
APP
|
amyloid beta (A4) precursor protein |
| chr12_-_76377795 | 1.06 |
ENST00000552856.1
|
RP11-114H23.1
|
RP11-114H23.1 |
| chr1_-_54355430 | 1.00 |
ENST00000371399.1
ENST00000072644.1 ENST00000412288.1 |
YIPF1
|
Yip1 domain family, member 1 |
| chr9_+_109685630 | 0.98 |
ENST00000451160.2
|
RP11-508N12.4
|
Uncharacterized protein |
| chr1_-_43919346 | 0.97 |
ENST00000372430.3
ENST00000372433.1 ENST00000372434.1 ENST00000486909.1 |
HYI
|
hydroxypyruvate isomerase (putative) |
| chr1_+_52682052 | 0.97 |
ENST00000371591.1
|
ZFYVE9
|
zinc finger, FYVE domain containing 9 |
| chr3_+_185046676 | 0.90 |
ENST00000428617.1
ENST00000443863.1 |
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr6_-_138833630 | 0.86 |
ENST00000533765.1
|
NHSL1
|
NHS-like 1 |
| chr5_-_148442584 | 0.86 |
ENST00000394358.2
ENST00000512049.1 |
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr8_-_37411648 | 0.86 |
ENST00000519738.1
|
RP11-150O12.1
|
RP11-150O12.1 |
| chr17_+_61086917 | 0.85 |
ENST00000424789.2
ENST00000389520.4 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr14_-_60952739 | 0.84 |
ENST00000555476.1
ENST00000321731.3 |
C14orf39
|
chromosome 14 open reading frame 39 |
| chr2_+_210444298 | 0.82 |
ENST00000445941.1
|
MAP2
|
microtubule-associated protein 2 |
| chr3_+_41241596 | 0.82 |
ENST00000450969.1
|
CTNNB1
|
catenin (cadherin-associated protein), beta 1, 88kDa |
| chr2_+_219081817 | 0.80 |
ENST00000315717.5
ENST00000420104.1 ENST00000295685.10 |
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr1_+_46152886 | 0.77 |
ENST00000372025.4
|
TMEM69
|
transmembrane protein 69 |
| chr6_+_47666275 | 0.75 |
ENST00000327753.3
ENST00000283303.2 |
GPR115
|
G protein-coupled receptor 115 |
| chr5_+_86564739 | 0.75 |
ENST00000456692.2
ENST00000512763.1 ENST00000506290.1 |
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1 |
| chr1_+_12524965 | 0.75 |
ENST00000471923.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr12_+_25205568 | 0.74 |
ENST00000548766.1
ENST00000556887.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr7_+_138915102 | 0.74 |
ENST00000486663.1
|
UBN2
|
ubinuclein 2 |
| chr3_+_63953415 | 0.73 |
ENST00000484332.1
|
ATXN7
|
ataxin 7 |
| chr4_+_174089951 | 0.73 |
ENST00000512285.1
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr21_-_27423339 | 0.72 |
ENST00000415997.1
|
APP
|
amyloid beta (A4) precursor protein |
| chr1_-_23495340 | 0.72 |
ENST00000418342.1
|
LUZP1
|
leucine zipper protein 1 |
| chr18_-_5396271 | 0.71 |
ENST00000579951.1
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr6_+_143447322 | 0.70 |
ENST00000458219.1
|
AIG1
|
androgen-induced 1 |
| chr8_-_73793975 | 0.68 |
ENST00000523881.1
|
RP11-1145L24.1
|
RP11-1145L24.1 |
| chr12_+_4130143 | 0.68 |
ENST00000543206.1
|
RP11-320N7.2
|
RP11-320N7.2 |
| chr12_+_14561422 | 0.68 |
ENST00000541056.1
|
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr1_+_109419804 | 0.66 |
ENST00000435475.1
|
GPSM2
|
G-protein signaling modulator 2 |
| chr17_-_40288449 | 0.66 |
ENST00000552162.1
ENST00000550504.1 |
RAB5C
|
RAB5C, member RAS oncogene family |
| chr11_+_118938485 | 0.65 |
ENST00000300793.6
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae) |
| chr2_+_138721850 | 0.65 |
ENST00000329366.4
ENST00000280097.3 |
HNMT
|
histamine N-methyltransferase |
| chr2_-_8977714 | 0.62 |
ENST00000319688.5
ENST00000489024.1 ENST00000256707.3 ENST00000427284.1 ENST00000418530.1 ENST00000473731.1 |
KIDINS220
|
kinase D-interacting substrate, 220kDa |
| chr10_+_76871353 | 0.62 |
ENST00000542569.1
|
SAMD8
|
sterile alpha motif domain containing 8 |
| chr4_+_26165074 | 0.61 |
ENST00000512351.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr2_+_233415363 | 0.58 |
ENST00000409514.1
ENST00000409098.1 ENST00000409495.1 ENST00000409167.3 ENST00000409322.1 ENST00000409394.1 |
EIF4E2
|
eukaryotic translation initiation factor 4E family member 2 |
| chr10_+_76871229 | 0.58 |
ENST00000372690.3
|
SAMD8
|
sterile alpha motif domain containing 8 |
| chr18_+_21032781 | 0.58 |
ENST00000339486.3
|
RIOK3
|
RIO kinase 3 |
| chr1_+_109419596 | 0.56 |
ENST00000435987.1
ENST00000264126.3 |
GPSM2
|
G-protein signaling modulator 2 |
| chr5_+_135468516 | 0.55 |
ENST00000507118.1
ENST00000511116.1 ENST00000545279.1 ENST00000545620.1 |
SMAD5
|
SMAD family member 5 |
| chr8_-_71157595 | 0.55 |
ENST00000519724.1
|
NCOA2
|
nuclear receptor coactivator 2 |
| chr12_+_32687221 | 0.55 |
ENST00000525053.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr5_+_36608280 | 0.55 |
ENST00000513646.1
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr1_+_89246647 | 0.54 |
ENST00000544045.1
|
PKN2
|
protein kinase N2 |
| chr6_+_158733692 | 0.54 |
ENST00000367094.2
ENST00000367097.3 |
TULP4
|
tubby like protein 4 |
| chr1_+_154401791 | 0.54 |
ENST00000476006.1
|
IL6R
|
interleukin 6 receptor |
| chr4_-_76957214 | 0.53 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chr19_+_21265028 | 0.53 |
ENST00000291770.7
|
ZNF714
|
zinc finger protein 714 |
| chr2_-_122262593 | 0.52 |
ENST00000418989.1
|
CLASP1
|
cytoplasmic linker associated protein 1 |
| chr10_+_85933494 | 0.52 |
ENST00000372126.3
|
C10orf99
|
chromosome 10 open reading frame 99 |
| chr7_+_134464414 | 0.51 |
ENST00000361901.2
|
CALD1
|
caldesmon 1 |
| chr7_+_57509877 | 0.51 |
ENST00000420713.1
|
ZNF716
|
zinc finger protein 716 |
| chr3_+_57875738 | 0.50 |
ENST00000417128.1
ENST00000438794.1 |
SLMAP
|
sarcolemma associated protein |
| chr6_-_26189304 | 0.49 |
ENST00000340756.2
|
HIST1H4D
|
histone cluster 1, H4d |
| chr3_+_171757346 | 0.49 |
ENST00000421757.1
ENST00000415807.2 ENST00000392699.1 |
FNDC3B
|
fibronectin type III domain containing 3B |
| chr4_-_39979576 | 0.48 |
ENST00000303538.8
ENST00000503396.1 |
PDS5A
|
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
| chr11_+_63606373 | 0.47 |
ENST00000402010.2
ENST00000315032.8 ENST00000377809.4 ENST00000413835.2 ENST00000377810.3 |
MARK2
|
MAP/microtubule affinity-regulating kinase 2 |
| chr2_+_151485403 | 0.46 |
ENST00000413247.1
ENST00000423428.1 ENST00000427615.1 ENST00000443811.1 |
AC104777.4
|
AC104777.4 |
| chr3_+_57875711 | 0.44 |
ENST00000442599.2
|
SLMAP
|
sarcolemma associated protein |
| chr10_+_13652047 | 0.44 |
ENST00000601460.1
|
RP11-295P9.3
|
Uncharacterized protein |
| chr12_+_19389814 | 0.44 |
ENST00000536974.1
|
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr4_+_169633310 | 0.43 |
ENST00000510998.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr5_-_16738451 | 0.43 |
ENST00000274203.9
ENST00000515803.1 |
MYO10
|
myosin X |
| chr7_-_107204918 | 0.42 |
ENST00000297135.3
|
COG5
|
component of oligomeric golgi complex 5 |
| chr16_-_15472151 | 0.41 |
ENST00000360151.4
ENST00000543801.1 |
NPIPA5
|
nuclear pore complex interacting protein family, member A5 |
| chr13_-_103719196 | 0.41 |
ENST00000245312.3
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter), member 2 |
| chr19_+_21264980 | 0.41 |
ENST00000596053.1
ENST00000597086.1 ENST00000596143.1 ENST00000596367.1 ENST00000601416.1 |
ZNF714
|
zinc finger protein 714 |
| chr22_+_32455111 | 0.41 |
ENST00000543737.1
|
SLC5A1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr15_+_22736246 | 0.41 |
ENST00000316397.3
|
GOLGA6L1
|
golgin A6 family-like 1 |
| chr16_-_25122735 | 0.40 |
ENST00000563176.1
|
RP11-449H11.1
|
RP11-449H11.1 |
| chr15_+_38746307 | 0.40 |
ENST00000397609.2
ENST00000491535.1 |
FAM98B
|
family with sequence similarity 98, member B |
| chr5_-_159846066 | 0.40 |
ENST00000519349.1
ENST00000520664.1 |
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae) |
| chr4_-_169239921 | 0.40 |
ENST00000514995.1
ENST00000393743.3 |
DDX60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr4_+_174089904 | 0.39 |
ENST00000265000.4
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr13_+_25338290 | 0.38 |
ENST00000255324.5
ENST00000381921.1 ENST00000255325.6 |
RNF17
|
ring finger protein 17 |
| chr4_-_48683188 | 0.37 |
ENST00000505759.1
|
FRYL
|
FRY-like |
| chr10_-_96829246 | 0.37 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr9_+_2029019 | 0.37 |
ENST00000382194.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr4_-_76944621 | 0.37 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr4_-_72649763 | 0.36 |
ENST00000513476.1
|
GC
|
group-specific component (vitamin D binding protein) |
| chr20_+_56964169 | 0.36 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr3_+_157827841 | 0.36 |
ENST00000295930.3
ENST00000471994.1 ENST00000464171.1 ENST00000312179.6 ENST00000475278.2 |
RSRC1
|
arginine/serine-rich coiled-coil 1 |
| chr6_-_133035185 | 0.35 |
ENST00000367928.4
|
VNN1
|
vanin 1 |
| chr14_+_52456327 | 0.35 |
ENST00000556760.1
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr19_+_21579958 | 0.35 |
ENST00000339914.6
ENST00000599461.1 |
ZNF493
|
zinc finger protein 493 |
| chr16_-_21431078 | 0.34 |
ENST00000458643.2
|
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr18_-_53177984 | 0.34 |
ENST00000543082.1
|
TCF4
|
transcription factor 4 |
| chr6_+_12958137 | 0.33 |
ENST00000457702.2
ENST00000379345.2 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr1_+_53480598 | 0.33 |
ENST00000430330.2
ENST00000408941.3 ENST00000478274.2 ENST00000484100.1 ENST00000435345.2 ENST00000488965.1 |
SCP2
|
sterol carrier protein 2 |
| chr1_+_151739131 | 0.33 |
ENST00000400999.1
|
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr2_-_42180940 | 0.33 |
ENST00000378711.2
|
C2orf91
|
chromosome 2 open reading frame 91 |
| chr18_-_52989217 | 0.33 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr6_+_125304502 | 0.33 |
ENST00000519799.1
ENST00000368414.2 ENST00000359704.2 |
RNF217
|
ring finger protein 217 |
| chr15_-_41836441 | 0.32 |
ENST00000567866.1
ENST00000561603.1 ENST00000304330.4 ENST00000566863.1 |
RPAP1
|
RNA polymerase II associated protein 1 |
| chr1_-_86848760 | 0.31 |
ENST00000460698.2
|
ODF2L
|
outer dense fiber of sperm tails 2-like |
| chr7_-_104567066 | 0.31 |
ENST00000453666.1
|
RP11-325F22.5
|
RP11-325F22.5 |
| chr2_+_208423891 | 0.31 |
ENST00000448277.1
ENST00000457101.1 |
CREB1
|
cAMP responsive element binding protein 1 |
| chr14_-_69444957 | 0.29 |
ENST00000556571.1
ENST00000553659.1 ENST00000555616.1 |
ACTN1
|
actinin, alpha 1 |
| chr2_-_169887827 | 0.29 |
ENST00000263817.6
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr16_+_14805546 | 0.28 |
ENST00000552140.1
|
NPIPA3
|
nuclear pore complex interacting protein family, member A3 |
| chr5_-_176433565 | 0.28 |
ENST00000428382.2
|
UIMC1
|
ubiquitin interaction motif containing 1 |
| chr1_+_210001309 | 0.27 |
ENST00000491415.2
|
DIEXF
|
digestive organ expansion factor homolog (zebrafish) |
| chr9_+_131062367 | 0.27 |
ENST00000601297.1
|
AL359091.2
|
CDNA: FLJ21673 fis, clone COL09042; HCG2036511; Uncharacterized protein |
| chr12_+_45686457 | 0.27 |
ENST00000441606.2
|
ANO6
|
anoctamin 6 |
| chr10_+_75504105 | 0.25 |
ENST00000535742.1
ENST00000546025.1 ENST00000345254.4 ENST00000540668.1 ENST00000339365.2 ENST00000411652.2 |
SEC24C
|
SEC24 family member C |
| chr14_-_50506589 | 0.25 |
ENST00000553914.2
|
RP11-58E21.3
|
RP11-58E21.3 |
| chr19_+_21264943 | 0.24 |
ENST00000597424.1
|
ZNF714
|
zinc finger protein 714 |
| chr15_+_22736484 | 0.23 |
ENST00000560659.2
|
GOLGA6L1
|
golgin A6 family-like 1 |
| chr19_-_23869999 | 0.23 |
ENST00000601935.1
ENST00000359788.4 ENST00000600313.1 ENST00000596211.1 ENST00000599168.1 |
ZNF675
|
zinc finger protein 675 |
| chr12_-_76879852 | 0.23 |
ENST00000548341.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr7_+_6121296 | 0.23 |
ENST00000428901.1
|
AC004895.4
|
AC004895.4 |
| chr1_+_178694300 | 0.22 |
ENST00000367635.3
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr14_+_52456193 | 0.22 |
ENST00000261700.3
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr10_-_129691195 | 0.22 |
ENST00000368671.3
|
CLRN3
|
clarin 3 |
| chr5_-_145562147 | 0.22 |
ENST00000545646.1
ENST00000274562.9 ENST00000510191.1 ENST00000394434.2 |
LARS
|
leucyl-tRNA synthetase |
| chr5_-_111093167 | 0.21 |
ENST00000446294.2
ENST00000419114.2 |
NREP
|
neuronal regeneration related protein |
| chr16_-_75498553 | 0.21 |
ENST00000569276.1
ENST00000357613.4 ENST00000561878.1 ENST00000566980.1 ENST00000567194.1 |
TMEM170A
RP11-77K12.1
|
transmembrane protein 170A Uncharacterized protein |
| chr1_-_204135450 | 0.20 |
ENST00000272190.8
ENST00000367195.2 |
REN
|
renin |
| chrX_+_117957741 | 0.20 |
ENST00000310164.2
|
ZCCHC12
|
zinc finger, CCHC domain containing 12 |
| chr19_+_42041860 | 0.19 |
ENST00000483481.2
ENST00000494375.2 |
AC006129.4
|
AC006129.4 |
| chr6_+_27782788 | 0.19 |
ENST00000359465.4
|
HIST1H2BM
|
histone cluster 1, H2bm |
| chr2_+_208423840 | 0.19 |
ENST00000539789.1
|
CREB1
|
cAMP responsive element binding protein 1 |
| chr18_+_3252265 | 0.19 |
ENST00000580887.1
ENST00000536605.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr1_-_237167718 | 0.18 |
ENST00000464121.2
|
MT1HL1
|
metallothionein 1H-like 1 |
| chr11_+_22688615 | 0.18 |
ENST00000533363.1
|
GAS2
|
growth arrest-specific 2 |
| chr5_-_138534071 | 0.18 |
ENST00000394817.2
|
SIL1
|
SIL1 nucleotide exchange factor |
| chr14_-_38028689 | 0.18 |
ENST00000553425.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr17_+_7211656 | 0.17 |
ENST00000416016.2
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr3_+_15045419 | 0.17 |
ENST00000406272.2
|
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr11_+_22688150 | 0.17 |
ENST00000454584.2
|
GAS2
|
growth arrest-specific 2 |
| chr11_-_60674000 | 0.17 |
ENST00000546152.1
|
PRPF19
|
pre-mRNA processing factor 19 |
| chr20_-_7921090 | 0.17 |
ENST00000378789.3
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr5_+_98109322 | 0.16 |
ENST00000513185.1
|
RGMB
|
repulsive guidance molecule family member b |
| chr16_-_25122785 | 0.15 |
ENST00000563962.1
ENST00000569920.1 |
RP11-449H11.1
|
RP11-449H11.1 |
| chr6_+_53948328 | 0.15 |
ENST00000370876.2
|
MLIP
|
muscular LMNA-interacting protein |
| chr4_-_186317034 | 0.14 |
ENST00000505916.1
|
LRP2BP
|
LRP2 binding protein |
| chr4_+_146539415 | 0.14 |
ENST00000281317.5
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr2_-_151428735 | 0.14 |
ENST00000441356.1
|
AC104777.2
|
AC104777.2 |
| chr12_-_62586543 | 0.14 |
ENST00000416284.3
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chr7_+_35756400 | 0.14 |
ENST00000585952.1
|
AC018647.3
|
AC018647.3 |
| chr2_-_39414848 | 0.13 |
ENST00000451199.1
|
CDKL4
|
cyclin-dependent kinase-like 4 |
| chr20_+_30102231 | 0.12 |
ENST00000335574.5
ENST00000340852.5 ENST00000398174.3 ENST00000376127.3 ENST00000344042.5 |
HM13
|
histocompatibility (minor) 13 |
| chr5_+_36606700 | 0.12 |
ENST00000416645.2
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr15_-_98417780 | 0.12 |
ENST00000503874.3
|
LINC00923
|
long intergenic non-protein coding RNA 923 |
| chr1_-_85100703 | 0.12 |
ENST00000370624.1
|
C1orf180
|
chromosome 1 open reading frame 180 |
| chr6_-_52859968 | 0.11 |
ENST00000370959.1
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr7_+_129984630 | 0.10 |
ENST00000355388.3
ENST00000497503.1 ENST00000463587.1 ENST00000461828.1 ENST00000494311.1 ENST00000466363.2 ENST00000485477.1 ENST00000431780.2 ENST00000474905.1 |
CPA5
|
carboxypeptidase A5 |
| chr7_+_93690435 | 0.10 |
ENST00000438538.1
|
AC003092.1
|
AC003092.1 |
| chr11_+_120039685 | 0.10 |
ENST00000530303.1
ENST00000319763.1 |
AP000679.2
|
Uncharacterized protein |
| chr16_-_4524543 | 0.09 |
ENST00000573571.1
ENST00000404295.3 ENST00000574425.1 |
NMRAL1
|
NmrA-like family domain containing 1 |
| chr14_+_73706308 | 0.09 |
ENST00000554301.1
ENST00000555445.1 |
PAPLN
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr14_+_53173910 | 0.08 |
ENST00000606149.1
ENST00000555339.1 ENST00000556813.1 |
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr5_+_177457525 | 0.08 |
ENST00000511856.1
ENST00000511189.1 |
FAM153C
|
family with sequence similarity 153, member C |
| chrX_-_131353461 | 0.08 |
ENST00000370874.1
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr8_+_107738240 | 0.08 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CDX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 1.0 | 3.0 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.9 | 2.7 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.4 | 4.3 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.4 | 5.3 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.4 | 1.9 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.4 | 1.1 | GO:0043317 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) regulation of cytotoxic T cell degranulation(GO:0043317) negative regulation of cytotoxic T cell degranulation(GO:0043318) |
| 0.4 | 1.8 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.3 | 1.3 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.2 | 1.2 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.2 | 0.7 | GO:0021503 | neural fold bending(GO:0021503) |
| 0.2 | 2.0 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.2 | 4.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.2 | 0.6 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.2 | 1.3 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.1 | 1.6 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.1 | 0.5 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.1 | 3.4 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.1 | 3.0 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.9 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.6 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.1 | 0.6 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.1 | 1.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.4 | GO:0039533 | regulation of MDA-5 signaling pathway(GO:0039533) |
| 0.1 | 0.6 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.1 | 6.5 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.1 | 0.9 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 0.7 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 0.6 | GO:1901189 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 0.1 | GO:0046666 | retinal cell programmed cell death(GO:0046666) regulation of retinal cell programmed cell death(GO:0046668) |
| 0.1 | 0.7 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 0.2 | GO:0006429 | glutaminyl-tRNA aminoacylation(GO:0006425) leucyl-tRNA aminoacylation(GO:0006429) |
| 0.1 | 0.4 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.5 | GO:0032916 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.1 | 0.3 | GO:1902304 | phosphatidylserine exposure on blood platelet(GO:0097045) positive regulation of potassium ion export(GO:1902304) |
| 0.1 | 0.7 | GO:0070777 | gamma-aminobutyric acid biosynthetic process(GO:0009449) D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 0.6 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.1 | 0.4 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.1 | 0.9 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 0.4 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 2.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.3 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.9 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.3 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 1.0 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.7 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.6 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.1 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.2 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 4.2 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.4 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.3 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.2 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.8 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 2.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 2.3 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.0 | 0.4 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.2 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.2 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 1.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 1.0 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.0 | 3.2 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 2.1 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.4 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.2 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.9 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.0 | 1.4 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 0.2 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.5 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.5 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.4 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.4 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.5 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.4 | GO:0007286 | spermatid development(GO:0007286) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.4 | 3.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.3 | 0.8 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.2 | 1.8 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.1 | 3.9 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.5 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.1 | 2.0 | GO:0097025 | lateral loop(GO:0043219) MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 1.6 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.5 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.8 | GO:0070369 | Scrib-APC-beta-catenin complex(GO:0034750) beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 1.0 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 3.0 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 0.5 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 0.5 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 3.7 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 0.6 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.6 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.9 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 2.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 1.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.6 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.7 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.8 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.6 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.3 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 1.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 1.8 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.9 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 1.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 5.0 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 2.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.3 | GO:0071141 | SMAD protein complex(GO:0071141) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.9 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.4 | 2.7 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 0.9 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.2 | 0.5 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.2 | 2.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 1.2 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 2.0 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 1.3 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 1.8 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 0.4 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.1 | 0.4 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.3 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.1 | 0.6 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 4.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 2.6 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 0.3 | GO:0015432 | bile acid-exporting ATPase activity(GO:0015432) |
| 0.1 | 3.5 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 2.7 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.4 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 0.4 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.1 | 0.2 | GO:0004823 | glutamine-tRNA ligase activity(GO:0004819) leucine-tRNA ligase activity(GO:0004823) |
| 0.1 | 0.4 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.7 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 1.0 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 1.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.4 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.2 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.1 | 3.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.5 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 1.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.8 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.6 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 2.6 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 2.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.4 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.3 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 1.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.7 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 1.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.6 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 6.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 1.0 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.6 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.3 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 2.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.5 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 1.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.6 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 4.1 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 5.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 3.6 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 6.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 2.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 4.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 2.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.5 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.8 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 1.9 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.9 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.7 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.7 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 3.6 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 2.0 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.9 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 5.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 4.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.4 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.6 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.4 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.5 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |