Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
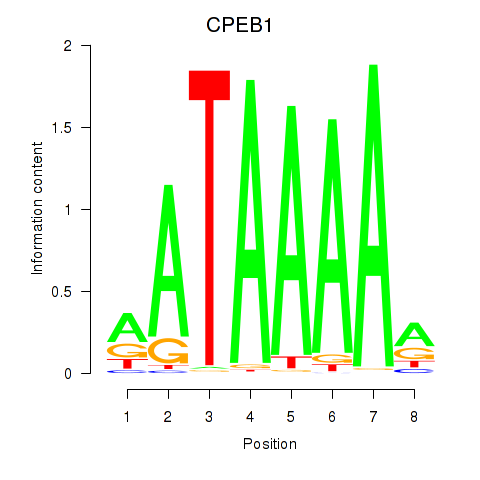
Results for CPEB1
Z-value: 1.18
Transcription factors associated with CPEB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CPEB1
|
ENSG00000214575.5 | cytoplasmic polyadenylation element binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CPEB1 | hg19_v2_chr15_-_83240553_83240576 | 0.91 | 4.1e-08 | Click! |
Activity profile of CPEB1 motif
Sorted Z-values of CPEB1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_32582293 | 5.62 |
ENST00000580907.1
ENST00000225831.4 |
CCL2
|
chemokine (C-C motif) ligand 2 |
| chr4_-_74864386 | 5.23 |
ENST00000296027.4
|
CXCL5
|
chemokine (C-X-C motif) ligand 5 |
| chr2_-_228244013 | 3.71 |
ENST00000304568.3
|
TM4SF20
|
transmembrane 4 L six family member 20 |
| chr6_+_62284008 | 3.56 |
ENST00000544932.1
|
MTRNR2L9
|
MT-RNR2-like 9 (pseudogene) |
| chr2_+_8822113 | 3.51 |
ENST00000396290.1
ENST00000331129.3 |
ID2
|
inhibitor of DNA binding 2, dominant negative helix-loop-helix protein |
| chr13_-_67802549 | 3.49 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr15_+_66994561 | 3.15 |
ENST00000288840.5
|
SMAD6
|
SMAD family member 6 |
| chr17_-_46806540 | 3.12 |
ENST00000290295.7
|
HOXB13
|
homeobox B13 |
| chr12_+_52203789 | 3.05 |
ENST00000599343.1
|
AC068987.1
|
HCG1997999; cDNA FLJ33996 fis, clone DFNES2008881 |
| chr12_+_54410664 | 2.89 |
ENST00000303406.4
|
HOXC4
|
homeobox C4 |
| chr6_+_12290586 | 2.84 |
ENST00000379375.5
|
EDN1
|
endothelin 1 |
| chr12_-_52715179 | 2.80 |
ENST00000293670.3
|
KRT83
|
keratin 83 |
| chr12_-_54071181 | 2.79 |
ENST00000338662.5
|
ATP5G2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
| chr6_-_64029879 | 2.76 |
ENST00000370658.5
ENST00000485906.2 ENST00000370657.4 |
LGSN
|
lengsin, lens protein with glutamine synthetase domain |
| chr8_-_145754428 | 2.72 |
ENST00000527462.1
ENST00000313465.5 ENST00000524821.1 |
C8orf82
|
chromosome 8 open reading frame 82 |
| chr2_-_190044480 | 2.61 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr5_+_92919043 | 2.53 |
ENST00000327111.3
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr17_+_47074758 | 2.53 |
ENST00000290341.3
|
IGF2BP1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr8_-_95220775 | 2.48 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr12_+_41221794 | 2.44 |
ENST00000547849.1
|
CNTN1
|
contactin 1 |
| chr1_-_238108575 | 2.41 |
ENST00000604646.1
|
MTRNR2L11
|
MT-RNR2-like 11 (pseudogene) |
| chr8_-_61193947 | 2.41 |
ENST00000317995.4
|
CA8
|
carbonic anhydrase VIII |
| chr3_-_155394099 | 2.40 |
ENST00000414191.1
|
PLCH1
|
phospholipase C, eta 1 |
| chr17_-_38574169 | 2.39 |
ENST00000423485.1
|
TOP2A
|
topoisomerase (DNA) II alpha 170kDa |
| chr1_+_164528866 | 2.35 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr17_-_47045949 | 2.31 |
ENST00000357424.2
|
GIP
|
gastric inhibitory polypeptide |
| chr6_+_127898312 | 2.28 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr11_-_27723158 | 2.27 |
ENST00000395980.2
|
BDNF
|
brain-derived neurotrophic factor |
| chr17_+_47075023 | 2.23 |
ENST00000431824.2
|
IGF2BP1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr19_+_36036477 | 2.22 |
ENST00000222284.5
ENST00000392204.2 |
TMEM147
|
transmembrane protein 147 |
| chr7_-_11871815 | 2.20 |
ENST00000423059.4
|
THSD7A
|
thrombospondin, type I, domain containing 7A |
| chr3_+_28283069 | 2.19 |
ENST00000466830.1
ENST00000423894.1 |
CMC1
|
C-x(9)-C motif containing 1 |
| chr18_-_25616519 | 2.18 |
ENST00000399380.3
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr3_-_155394152 | 2.18 |
ENST00000494598.1
|
PLCH1
|
phospholipase C, eta 1 |
| chr8_-_95274536 | 2.09 |
ENST00000297596.2
ENST00000396194.2 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr15_-_56209306 | 2.08 |
ENST00000506154.1
ENST00000338963.2 ENST00000508342.1 |
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
| chr11_+_123301012 | 2.04 |
ENST00000533341.1
|
AP000783.1
|
Uncharacterized protein |
| chr5_-_58295712 | 2.03 |
ENST00000317118.8
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr10_+_94590910 | 2.03 |
ENST00000371547.4
|
EXOC6
|
exocyst complex component 6 |
| chr10_-_21806759 | 2.01 |
ENST00000444772.3
|
SKIDA1
|
SKI/DACH domain containing 1 |
| chr2_-_175711133 | 2.00 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr11_+_71934962 | 1.99 |
ENST00000543234.1
|
INPPL1
|
inositol polyphosphate phosphatase-like 1 |
| chr12_+_52695617 | 1.98 |
ENST00000293525.5
|
KRT86
|
keratin 86 |
| chr12_+_54393880 | 1.96 |
ENST00000303450.4
|
HOXC9
|
homeobox C9 |
| chr12_-_52761262 | 1.96 |
ENST00000257901.3
|
KRT85
|
keratin 85 |
| chr2_+_145780739 | 1.88 |
ENST00000597173.1
ENST00000602108.1 ENST00000420472.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr17_+_79369249 | 1.88 |
ENST00000574717.2
|
RP11-1055B8.6
|
Uncharacterized protein |
| chr19_+_36119975 | 1.85 |
ENST00000589559.1
ENST00000360475.4 |
RBM42
|
RNA binding motif protein 42 |
| chr6_+_111195973 | 1.85 |
ENST00000368885.3
ENST00000368882.3 ENST00000451850.2 ENST00000368877.5 |
AMD1
|
adenosylmethionine decarboxylase 1 |
| chr3_-_148939598 | 1.81 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr8_+_71485681 | 1.81 |
ENST00000391684.1
|
AC120194.1
|
AC120194.1 |
| chr1_+_164529004 | 1.80 |
ENST00000559240.1
ENST00000367897.1 ENST00000540236.1 ENST00000401534.1 |
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr5_-_59783882 | 1.80 |
ENST00000505507.2
ENST00000502484.2 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr10_-_69597810 | 1.79 |
ENST00000483798.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr11_-_10822029 | 1.78 |
ENST00000528839.1
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr3_-_148939835 | 1.78 |
ENST00000264613.6
|
CP
|
ceruloplasmin (ferroxidase) |
| chr8_-_38326139 | 1.77 |
ENST00000335922.5
ENST00000532791.1 ENST00000397091.5 |
FGFR1
|
fibroblast growth factor receptor 1 |
| chr17_-_78450398 | 1.74 |
ENST00000306773.4
|
NPTX1
|
neuronal pentraxin I |
| chr19_+_36036583 | 1.73 |
ENST00000392205.1
|
TMEM147
|
transmembrane protein 147 |
| chr12_+_54422142 | 1.72 |
ENST00000243108.4
|
HOXC6
|
homeobox C6 |
| chr11_-_115127611 | 1.70 |
ENST00000545094.1
|
CADM1
|
cell adhesion molecule 1 |
| chr6_-_127780510 | 1.67 |
ENST00000487331.2
ENST00000483725.3 |
KIAA0408
|
KIAA0408 |
| chr7_-_27219849 | 1.67 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr9_-_119162885 | 1.66 |
ENST00000445861.2
|
PAPPA-AS1
|
PAPPA antisense RNA 1 |
| chr9_-_20622478 | 1.65 |
ENST00000355930.6
ENST00000380338.4 |
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr10_-_69597828 | 1.63 |
ENST00000339758.7
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr11_-_115158193 | 1.61 |
ENST00000543540.1
|
CADM1
|
cell adhesion molecule 1 |
| chrX_-_100662881 | 1.58 |
ENST00000218516.3
|
GLA
|
galactosidase, alpha |
| chr7_-_33080506 | 1.58 |
ENST00000381626.2
ENST00000409467.1 ENST00000449201.1 |
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr7_+_66800928 | 1.58 |
ENST00000430244.1
|
RP11-166O4.5
|
RP11-166O4.5 |
| chr17_-_17480779 | 1.57 |
ENST00000395782.1
|
PEMT
|
phosphatidylethanolamine N-methyltransferase |
| chr2_+_152214098 | 1.57 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr8_+_77593448 | 1.56 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr17_-_76220740 | 1.54 |
ENST00000600484.1
|
AC087645.1
|
Uncharacterized protein |
| chr19_+_36120009 | 1.53 |
ENST00000589871.1
|
RBM42
|
RNA binding motif protein 42 |
| chr8_+_26435359 | 1.51 |
ENST00000311151.5
|
DPYSL2
|
dihydropyrimidinase-like 2 |
| chr2_-_148779106 | 1.51 |
ENST00000416719.1
ENST00000264169.2 |
ORC4
|
origin recognition complex, subunit 4 |
| chrX_+_72783026 | 1.51 |
ENST00000373504.6
ENST00000373502.5 |
CHIC1
|
cysteine-rich hydrophobic domain 1 |
| chrX_-_20236970 | 1.50 |
ENST00000379548.4
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr16_-_2314222 | 1.50 |
ENST00000566397.1
|
RNPS1
|
RNA binding protein S1, serine-rich domain |
| chrX_-_20237059 | 1.50 |
ENST00000457145.1
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr2_+_176981307 | 1.50 |
ENST00000249501.4
|
HOXD10
|
homeobox D10 |
| chr8_+_26435915 | 1.48 |
ENST00000523027.1
|
DPYSL2
|
dihydropyrimidinase-like 2 |
| chr17_-_2117600 | 1.45 |
ENST00000572369.1
|
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr1_-_32384693 | 1.45 |
ENST00000602683.1
ENST00000470404.1 |
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2 |
| chr1_+_244214577 | 1.45 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr7_-_14029515 | 1.43 |
ENST00000430479.1
ENST00000405218.2 ENST00000343495.5 |
ETV1
|
ets variant 1 |
| chr19_+_36119929 | 1.43 |
ENST00000588161.1
ENST00000262633.4 ENST00000592202.1 ENST00000586618.1 |
RBM42
|
RNA binding motif protein 42 |
| chr6_-_41909191 | 1.43 |
ENST00000512426.1
ENST00000372987.4 |
CCND3
|
cyclin D3 |
| chrX_-_55208866 | 1.42 |
ENST00000545075.1
|
MTRNR2L10
|
MT-RNR2-like 10 |
| chr5_+_32788945 | 1.41 |
ENST00000326958.1
|
AC026703.1
|
AC026703.1 |
| chr12_+_85673868 | 1.39 |
ENST00000316824.3
|
ALX1
|
ALX homeobox 1 |
| chr8_-_38326119 | 1.38 |
ENST00000356207.5
ENST00000326324.6 |
FGFR1
|
fibroblast growth factor receptor 1 |
| chr18_+_3466248 | 1.37 |
ENST00000581029.1
ENST00000581442.1 ENST00000579007.1 |
RP11-838N2.4
|
RP11-838N2.4 |
| chr12_+_58003935 | 1.34 |
ENST00000333972.7
|
ARHGEF25
|
Rho guanine nucleotide exchange factor (GEF) 25 |
| chr2_+_176987088 | 1.34 |
ENST00000249499.6
|
HOXD9
|
homeobox D9 |
| chr18_+_68002675 | 1.33 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr17_+_59477233 | 1.33 |
ENST00000240328.3
|
TBX2
|
T-box 2 |
| chr2_-_74726710 | 1.25 |
ENST00000377566.4
|
LBX2
|
ladybird homeobox 2 |
| chr1_+_164528616 | 1.24 |
ENST00000340699.3
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr1_+_200011711 | 1.22 |
ENST00000544748.1
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr16_+_11439286 | 1.22 |
ENST00000312499.5
ENST00000576027.1 |
RMI2
|
RecQ mediated genome instability 2 |
| chr1_+_212475148 | 1.22 |
ENST00000537030.3
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr20_+_42574317 | 1.21 |
ENST00000358131.5
|
TOX2
|
TOX high mobility group box family member 2 |
| chr19_+_50191921 | 1.20 |
ENST00000420022.3
|
ADM5
|
adrenomedullin 5 (putative) |
| chr17_+_36886478 | 1.18 |
ENST00000439660.2
|
CISD3
|
CDGSH iron sulfur domain 3 |
| chr2_+_145780767 | 1.17 |
ENST00000599358.1
ENST00000596278.1 ENST00000596747.1 ENST00000608652.1 ENST00000609705.1 ENST00000608432.1 ENST00000596970.1 ENST00000602041.1 ENST00000601578.1 ENST00000596034.1 ENST00000414195.2 ENST00000594837.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr16_+_19222479 | 1.17 |
ENST00000568433.1
|
SYT17
|
synaptotagmin XVII |
| chr17_-_76921459 | 1.14 |
ENST00000262768.7
|
TIMP2
|
TIMP metallopeptidase inhibitor 2 |
| chr9_+_97562440 | 1.13 |
ENST00000395357.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr10_+_17271266 | 1.11 |
ENST00000224237.5
|
VIM
|
vimentin |
| chr1_-_25291475 | 1.09 |
ENST00000338888.3
ENST00000399916.1 |
RUNX3
|
runt-related transcription factor 3 |
| chr8_-_25281747 | 1.08 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr2_+_114256661 | 1.07 |
ENST00000306507.5
|
FOXD4L1
|
forkhead box D4-like 1 |
| chr3_-_114035026 | 1.07 |
ENST00000570269.1
|
RP11-553L6.5
|
RP11-553L6.5 |
| chr9_-_16870704 | 1.02 |
ENST00000380672.4
ENST00000380667.2 ENST00000380666.2 ENST00000486514.1 |
BNC2
|
basonuclin 2 |
| chr9_-_133814527 | 1.00 |
ENST00000451466.1
|
FIBCD1
|
fibrinogen C domain containing 1 |
| chr6_+_44194762 | 1.00 |
ENST00000371708.1
|
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr14_+_56078695 | 1.00 |
ENST00000416613.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr3_+_158991025 | 0.98 |
ENST00000337808.6
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr1_+_241695670 | 0.97 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr17_-_73775839 | 0.97 |
ENST00000592643.1
ENST00000591890.1 ENST00000587171.1 ENST00000254810.4 ENST00000589599.1 |
H3F3B
|
H3 histone, family 3B (H3.3B) |
| chr4_+_169575875 | 0.96 |
ENST00000503457.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr3_+_142342228 | 0.96 |
ENST00000337777.3
|
PLS1
|
plastin 1 |
| chr7_+_95115210 | 0.95 |
ENST00000428113.1
ENST00000325885.5 |
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr5_+_135496675 | 0.94 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chr19_-_46272462 | 0.94 |
ENST00000317578.6
|
SIX5
|
SIX homeobox 5 |
| chr11_+_77300669 | 0.94 |
ENST00000313578.3
|
AQP11
|
aquaporin 11 |
| chr6_+_26104104 | 0.93 |
ENST00000377803.2
|
HIST1H4C
|
histone cluster 1, H4c |
| chr8_-_95449155 | 0.92 |
ENST00000481490.2
|
FSBP
|
fibrinogen silencer binding protein |
| chr2_-_145277569 | 0.92 |
ENST00000303660.4
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr8_+_77593474 | 0.92 |
ENST00000455469.2
ENST00000050961.6 |
ZFHX4
|
zinc finger homeobox 4 |
| chr12_+_79258547 | 0.91 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr1_+_221051699 | 0.90 |
ENST00000366903.6
|
HLX
|
H2.0-like homeobox |
| chr8_+_94752349 | 0.90 |
ENST00000391680.1
|
RBM12B-AS1
|
RBM12B antisense RNA 1 |
| chr12_+_41221975 | 0.90 |
ENST00000552913.1
|
CNTN1
|
contactin 1 |
| chr17_-_53800217 | 0.88 |
ENST00000424486.2
|
TMEM100
|
transmembrane protein 100 |
| chrX_+_106871713 | 0.88 |
ENST00000372435.4
ENST00000372428.4 ENST00000372419.3 ENST00000543248.1 |
PRPS1
|
phosphoribosyl pyrophosphate synthetase 1 |
| chr12_-_118406028 | 0.87 |
ENST00000425217.1
|
KSR2
|
kinase suppressor of ras 2 |
| chr22_-_41258074 | 0.86 |
ENST00000307221.4
|
DNAJB7
|
DnaJ (Hsp40) homolog, subfamily B, member 7 |
| chr7_-_14029283 | 0.85 |
ENST00000433547.1
ENST00000405192.2 |
ETV1
|
ets variant 1 |
| chr2_-_148778323 | 0.85 |
ENST00000440042.1
ENST00000535373.1 ENST00000540442.1 ENST00000536575.1 |
ORC4
|
origin recognition complex, subunit 4 |
| chr11_-_67205538 | 0.84 |
ENST00000326294.3
|
PTPRCAP
|
protein tyrosine phosphatase, receptor type, C-associated protein |
| chr17_-_46667594 | 0.84 |
ENST00000476342.1
ENST00000460160.1 ENST00000472863.1 |
HOXB3
|
homeobox B3 |
| chr17_+_73780852 | 0.83 |
ENST00000589666.1
|
UNK
|
unkempt family zinc finger |
| chr6_+_26156551 | 0.83 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr4_-_105416039 | 0.83 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chrX_-_100604184 | 0.83 |
ENST00000372902.3
|
TIMM8A
|
translocase of inner mitochondrial membrane 8 homolog A (yeast) |
| chr2_-_55646412 | 0.82 |
ENST00000413716.2
|
CCDC88A
|
coiled-coil domain containing 88A |
| chrX_+_92929192 | 0.82 |
ENST00000332647.4
|
FAM133A
|
family with sequence similarity 133, member A |
| chr9_+_74920335 | 0.81 |
ENST00000451596.2
ENST00000436054.1 |
RP11-63P12.6
|
RP11-63P12.6 |
| chr9_-_139137648 | 0.81 |
ENST00000358701.5
|
QSOX2
|
quiescin Q6 sulfhydryl oxidase 2 |
| chr9_+_74920408 | 0.80 |
ENST00000451152.1
|
RP11-63P12.6
|
RP11-63P12.6 |
| chr1_+_95616933 | 0.80 |
ENST00000604203.1
|
RP11-57H12.6
|
TMEM56-RWDD3 readthrough |
| chr5_+_139505520 | 0.80 |
ENST00000333305.3
|
IGIP
|
IgA-inducing protein |
| chr3_+_148545586 | 0.80 |
ENST00000282957.4
ENST00000468341.1 |
CPB1
|
carboxypeptidase B1 (tissue) |
| chr7_+_7811992 | 0.80 |
ENST00000406829.1
|
RPA3-AS1
|
RPA3 antisense RNA 1 |
| chr3_+_12392971 | 0.80 |
ENST00000287820.6
|
PPARG
|
peroxisome proliferator-activated receptor gamma |
| chr12_+_79258444 | 0.79 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr1_-_230991747 | 0.78 |
ENST00000523410.1
|
C1orf198
|
chromosome 1 open reading frame 198 |
| chr11_+_123325106 | 0.78 |
ENST00000525757.1
|
LINC01059
|
long intergenic non-protein coding RNA 1059 |
| chr2_-_145277640 | 0.77 |
ENST00000539609.3
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chrX_-_77395186 | 0.77 |
ENST00000341864.5
|
TAF9B
|
TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa |
| chr6_+_10556215 | 0.77 |
ENST00000316170.3
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr9_+_116263778 | 0.76 |
ENST00000394646.3
|
RGS3
|
regulator of G-protein signaling 3 |
| chr2_+_145780725 | 0.76 |
ENST00000451478.1
|
TEX41
|
testis expressed 41 (non-protein coding) |
| chrX_+_123095860 | 0.76 |
ENST00000428941.1
|
STAG2
|
stromal antigen 2 |
| chrX_-_46187069 | 0.76 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr1_+_26798955 | 0.75 |
ENST00000361427.5
|
HMGN2
|
high mobility group nucleosomal binding domain 2 |
| chr6_+_100054606 | 0.75 |
ENST00000369215.4
|
PRDM13
|
PR domain containing 13 |
| chr12_+_72058130 | 0.75 |
ENST00000547843.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr9_-_133814455 | 0.74 |
ENST00000448616.1
|
FIBCD1
|
fibrinogen C domain containing 1 |
| chr1_-_103574024 | 0.74 |
ENST00000512756.1
ENST00000370096.3 ENST00000358392.2 ENST00000353414.4 |
COL11A1
|
collagen, type XI, alpha 1 |
| chr1_+_153190053 | 0.74 |
ENST00000368744.3
|
PRR9
|
proline rich 9 |
| chr4_-_74904398 | 0.74 |
ENST00000296026.4
|
CXCL3
|
chemokine (C-X-C motif) ligand 3 |
| chr2_+_183582774 | 0.73 |
ENST00000537515.1
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr17_-_56591978 | 0.73 |
ENST00000583656.1
|
MTMR4
|
myotubularin related protein 4 |
| chr9_-_69229650 | 0.72 |
ENST00000416428.1
|
CBWD6
|
COBW domain containing 6 |
| chr16_-_3422283 | 0.72 |
ENST00000399974.3
|
MTRNR2L4
|
MT-RNR2-like 4 |
| chr10_-_10504285 | 0.72 |
ENST00000602311.1
|
RP11-271F18.4
|
RP11-271F18.4 |
| chr9_+_135037334 | 0.72 |
ENST00000393229.3
ENST00000360670.3 ENST00000393228.4 ENST00000372179.3 |
NTNG2
|
netrin G2 |
| chr3_-_185270383 | 0.71 |
ENST00000296252.4
|
LIPH
|
lipase, member H |
| chr4_+_95972822 | 0.71 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr6_-_31697977 | 0.71 |
ENST00000375787.2
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chrX_-_154299501 | 0.70 |
ENST00000369476.3
ENST00000369484.3 |
MTCP1
CMC4
|
mature T-cell proliferation 1 C-x(9)-C motif containing 4 |
| chr3_-_185270342 | 0.70 |
ENST00000424591.2
|
LIPH
|
lipase, member H |
| chr6_+_33172407 | 0.70 |
ENST00000374662.3
|
HSD17B8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr2_+_67624430 | 0.69 |
ENST00000272342.5
|
ETAA1
|
Ewing tumor-associated antigen 1 |
| chr1_-_200379180 | 0.69 |
ENST00000294740.3
|
ZNF281
|
zinc finger protein 281 |
| chr6_-_152489484 | 0.69 |
ENST00000354674.4
ENST00000539504.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chrX_-_19689106 | 0.68 |
ENST00000379716.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr4_-_174451370 | 0.68 |
ENST00000359562.4
|
HAND2
|
heart and neural crest derivatives expressed 2 |
| chrX_-_54209640 | 0.68 |
ENST00000375180.2
ENST00000328235.4 ENST00000477084.1 |
FAM120C
|
family with sequence similarity 120C |
| chr17_+_42264556 | 0.68 |
ENST00000319511.6
ENST00000589785.1 ENST00000592825.1 ENST00000589184.1 |
TMUB2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr11_-_62473776 | 0.68 |
ENST00000278893.7
ENST00000407022.3 ENST00000421906.1 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr14_-_95236551 | 0.67 |
ENST00000238558.3
|
GSC
|
goosecoid homeobox |
| chr1_-_11918988 | 0.67 |
ENST00000376468.3
|
NPPB
|
natriuretic peptide B |
| chr1_+_84630053 | 0.66 |
ENST00000394838.2
ENST00000370682.3 ENST00000432111.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr2_+_207308220 | 0.66 |
ENST00000264377.3
|
ADAM23
|
ADAM metallopeptidase domain 23 |
| chr16_+_2303738 | 0.65 |
ENST00000454671.1
|
AC009065.1
|
Uncharacterized protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of CPEB1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.6 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 1.0 | 3.1 | GO:1903465 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.9 | 2.6 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.8 | 3.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.8 | 2.3 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.8 | 3.0 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.7 | 3.5 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.7 | 2.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.7 | 2.8 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.6 | 1.7 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.6 | 4.0 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.6 | 3.3 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.5 | 2.5 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.5 | 1.8 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.4 | 1.3 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.4 | 4.8 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.4 | 3.8 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.4 | 2.1 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.3 | 1.3 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.3 | 2.3 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.3 | 0.9 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.3 | 0.9 | GO:0046101 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.3 | 1.7 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.3 | 1.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.2 | 2.4 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.2 | 1.6 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.2 | 1.6 | GO:0016139 | glycoside catabolic process(GO:0016139) glycosylceramide catabolic process(GO:0046477) |
| 0.2 | 2.2 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.2 | 2.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.2 | 1.5 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.2 | 1.4 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.2 | 0.8 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.2 | 1.8 | GO:0061373 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.2 | 5.4 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.2 | 1.0 | GO:0072343 | pancreatic stellate cell proliferation(GO:0072343) regulation of pancreatic stellate cell proliferation(GO:2000229) |
| 0.2 | 1.0 | GO:0015862 | uridine transport(GO:0015862) |
| 0.2 | 5.8 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.2 | 1.1 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.2 | 1.5 | GO:0021546 | rhombomere development(GO:0021546) |
| 0.2 | 0.7 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.2 | 0.5 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.2 | 1.2 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.2 | 0.5 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.2 | 6.4 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.2 | 1.6 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.2 | 0.9 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.1 | 0.4 | GO:1903980 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) positive regulation of microglial cell activation(GO:1903980) |
| 0.1 | 6.3 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 0.8 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 2.1 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.1 | 0.4 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 | 0.5 | GO:0043375 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.1 | 0.5 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.1 | 1.9 | GO:0007379 | segment specification(GO:0007379) |
| 0.1 | 0.5 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.1 | 1.6 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.7 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.1 | 0.9 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 3.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 1.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.7 | GO:0035989 | tendon development(GO:0035989) |
| 0.1 | 0.7 | GO:1903288 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 0.7 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 0.9 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 0.3 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.1 | 0.8 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 1.3 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 0.3 | GO:1990641 | cellular response to bile acid(GO:1903413) negative regulation of intestinal absorption(GO:1904479) response to iron ion starvation(GO:1990641) |
| 0.1 | 0.7 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.5 | GO:0061325 | cell proliferation involved in outflow tract morphogenesis(GO:0061325) |
| 0.1 | 0.5 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.3 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.8 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.2 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 | 0.5 | GO:2000501 | regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.1 | 1.0 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 1.1 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 0.4 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 0.6 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 0.6 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.1 | 0.2 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.1 | 2.8 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 1.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.2 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.1 | 0.2 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.1 | 0.9 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 0.6 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.1 | 0.3 | GO:0045209 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.1 | 0.5 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 | 2.7 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.1 | 1.2 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.1 | 0.7 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 0.5 | GO:0002740 | negative regulation of cytokine secretion involved in immune response(GO:0002740) Tie signaling pathway(GO:0048014) |
| 0.1 | 0.4 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.1 | 4.0 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.1 | 0.7 | GO:0023019 | signal transduction involved in regulation of gene expression(GO:0023019) |
| 0.1 | 6.5 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.1 | 1.7 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.6 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 1.1 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.5 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.5 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.8 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.6 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.2 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 3.0 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 1.0 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 2.6 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.3 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.7 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:0048867 | ganglion mother cell fate determination(GO:0007402) stem cell fate determination(GO:0048867) |
| 0.0 | 1.0 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 0.2 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.7 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.3 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.3 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.4 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 1.2 | GO:0000732 | strand displacement(GO:0000732) |
| 0.0 | 0.7 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.3 | GO:0006091 | generation of precursor metabolites and energy(GO:0006091) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.5 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.2 | GO:0046034 | ATP metabolic process(GO:0046034) |
| 0.0 | 0.2 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.0 | 0.8 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.4 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.7 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.8 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.7 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 3.8 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 6.7 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 1.3 | GO:0031640 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.0 | 0.2 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.4 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 1.4 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.4 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:0008582 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 2.0 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 2.9 | GO:0042633 | molting cycle(GO:0042303) hair cycle(GO:0042633) |
| 0.0 | 0.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 1.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.3 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.2 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.2 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.2 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.0 | 0.1 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.5 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.6 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.2 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.6 | GO:0006625 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.2 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.2 | GO:0090656 | t-circle formation(GO:0090656) |
| 0.0 | 0.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.3 | GO:0031647 | regulation of protein stability(GO:0031647) |
| 0.0 | 0.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.4 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 1.4 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.8 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.2 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 0.5 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.1 | GO:1900046 | regulation of blood coagulation(GO:0030193) regulation of hemostasis(GO:1900046) |
| 0.0 | 0.2 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.0 | GO:0001812 | regulation of type I hypersensitivity(GO:0001810) positive regulation of type I hypersensitivity(GO:0001812) B cell cytokine production(GO:0002368) type I hypersensitivity(GO:0016068) |
| 0.0 | 0.2 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.2 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.2 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.3 | GO:0019054 | modulation by virus of host process(GO:0019054) |
| 0.0 | 0.3 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.8 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 0.1 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.8 | 2.4 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.7 | 2.8 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.3 | 4.8 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.3 | 1.5 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.2 | 0.2 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.2 | 0.9 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.2 | 1.7 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.2 | 3.0 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 1.7 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.2 | 1.5 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 2.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 0.9 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.4 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 1.3 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.5 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 1.0 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 2.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 1.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.6 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.7 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 2.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 3.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.9 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.8 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 1.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.3 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 2.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.7 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.2 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.1 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 1.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 3.0 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.8 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.9 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 1.9 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 1.4 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 2.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.5 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.4 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.2 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 4.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.5 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 3.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 2.1 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.7 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 1.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.3 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 1.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.5 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.2 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.9 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 1.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.3 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.9 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 2.4 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 2.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.8 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 2.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 1.1 | 3.2 | GO:0070698 | transforming growth factor beta receptor, inhibitory cytoplasmic mediator activity(GO:0030617) type I activin receptor binding(GO:0070698) |
| 0.8 | 2.5 | GO:0005427 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.8 | 2.4 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.8 | 2.3 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.7 | 2.8 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.6 | 1.8 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.6 | 3.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.6 | 2.8 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.5 | 1.6 | GO:0004608 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.4 | 1.4 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.3 | 1.6 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.3 | 4.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.3 | 0.9 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.3 | 3.6 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 5.9 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.3 | 3.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.3 | 2.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 0.7 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.2 | 0.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 1.6 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 5.9 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.2 | 1.0 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.2 | 1.7 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.2 | 1.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.2 | 0.8 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 2.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.8 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.1 | 3.0 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 2.0 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.1 | 4.8 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.3 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.1 | 0.3 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.1 | 0.7 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 3.0 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.5 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.5 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 1.7 | GO:0008061 | chitin binding(GO:0008061) |
| 0.1 | 0.8 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.6 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.2 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.1 | 0.7 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 2.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 1.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.5 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 0.3 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 0.7 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.1 | 0.5 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.1 | 3.0 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.4 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.6 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.9 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 1.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.0 | 0.7 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.8 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.3 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 5.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.4 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.3 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.8 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.4 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.3 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 1.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 1.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.5 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.7 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.9 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 1.5 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 2.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 1.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.2 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.0 | 2.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.6 | GO:0008430 | selenium binding(GO:0008430) inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 1.0 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.3 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.9 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 4.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.2 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.2 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 2.7 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 2.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.5 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.4 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 2.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.0 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.4 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 3.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 1.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.3 | GO:0030291 | protein serine/threonine kinase inhibitor activity(GO:0030291) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 1.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.6 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.3 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.6 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 1.4 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.3 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 2.7 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 4.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 7.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 0.9 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 2.5 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 2.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 3.1 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.8 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 1.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.7 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 4.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 2.8 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 2.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 4.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 2.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 2.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.8 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.8 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.8 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.7 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 2.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.2 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.4 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.7 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 1.2 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.0 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.2 | 2.8 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.2 | 11.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 1.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 0.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 6.7 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 4.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 2.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 2.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.7 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 1.8 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 2.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 3.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 3.5 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 6.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 1.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 1.5 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 1.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.5 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 2.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.7 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 4.4 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 3.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 2.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.6 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 MUTANTS | Genes involved in Signaling by FGFR1 mutants |
| 0.0 | 1.0 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 2.5 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.2 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 1.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 1.3 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 2.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |