Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
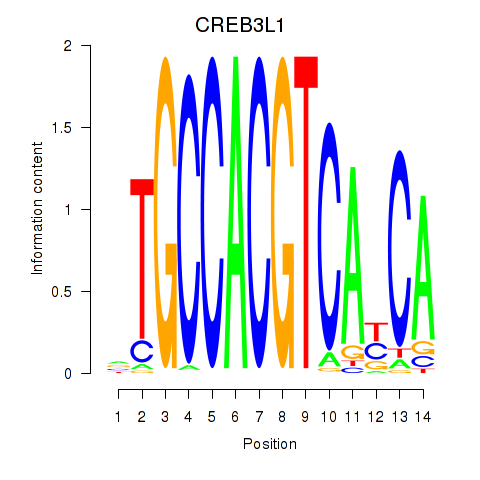
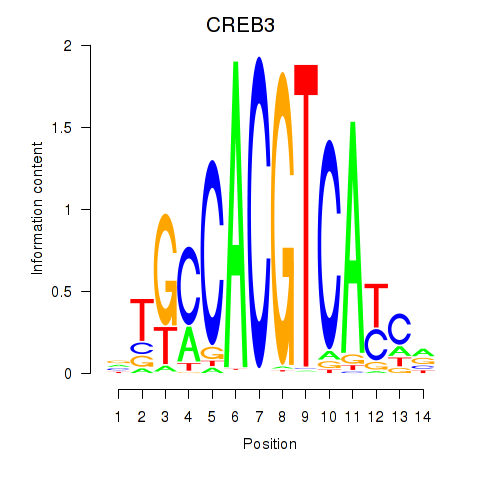
Results for CREB3L1_CREB3
Z-value: 0.26
Transcription factors associated with CREB3L1_CREB3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CREB3L1
|
ENSG00000157613.6 | cAMP responsive element binding protein 3 like 1 |
|
CREB3
|
ENSG00000107175.6 | cAMP responsive element binding protein 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CREB3L1 | hg19_v2_chr11_+_46316677_46316719 | 0.31 | 1.8e-01 | Click! |
| CREB3 | hg19_v2_chr9_+_35732312_35732332 | -0.03 | 8.9e-01 | Click! |
Activity profile of CREB3L1_CREB3 motif
Sorted Z-values of CREB3L1_CREB3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_116692102 | 0.69 |
ENST00000359564.2
|
DSE
|
dermatan sulfate epimerase |
| chr1_-_113498616 | 0.42 |
ENST00000433570.4
ENST00000538576.1 ENST00000458229.1 |
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr16_-_68269971 | 0.40 |
ENST00000565858.1
|
ESRP2
|
epithelial splicing regulatory protein 2 |
| chr1_-_113498943 | 0.36 |
ENST00000369626.3
|
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr3_+_50284321 | 0.35 |
ENST00000451956.1
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 |
| chr20_-_1306351 | 0.34 |
ENST00000381812.1
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr6_+_32937083 | 0.34 |
ENST00000456339.1
|
BRD2
|
bromodomain containing 2 |
| chr20_-_1306391 | 0.33 |
ENST00000339987.3
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr19_+_38810447 | 0.31 |
ENST00000263372.3
|
KCNK6
|
potassium channel, subfamily K, member 6 |
| chr7_+_128379449 | 0.31 |
ENST00000479257.1
|
CALU
|
calumenin |
| chr12_-_48276710 | 0.28 |
ENST00000550314.1
|
VDR
|
vitamin D (1,25- dihydroxyvitamin D3) receptor |
| chr10_-_27444143 | 0.27 |
ENST00000477432.1
|
YME1L1
|
YME1-like 1 ATPase |
| chr5_+_133984462 | 0.26 |
ENST00000398844.2
ENST00000322887.4 |
SEC24A
|
SEC24 family member A |
| chr9_+_124461603 | 0.25 |
ENST00000373782.3
|
DAB2IP
|
DAB2 interacting protein |
| chr3_+_100211412 | 0.25 |
ENST00000323523.4
ENST00000403410.1 ENST00000449609.1 |
TMEM45A
|
transmembrane protein 45A |
| chr4_+_75311019 | 0.23 |
ENST00000502307.1
|
AREG
|
amphiregulin |
| chr5_-_131563474 | 0.21 |
ENST00000417528.1
|
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chr13_-_52027134 | 0.21 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr11_-_59383617 | 0.21 |
ENST00000263847.1
|
OSBP
|
oxysterol binding protein |
| chr12_-_50616122 | 0.20 |
ENST00000552823.1
ENST00000552909.1 |
LIMA1
|
LIM domain and actin binding 1 |
| chr19_-_46916805 | 0.20 |
ENST00000307522.3
|
CCDC8
|
coiled-coil domain containing 8 |
| chr2_+_219745020 | 0.19 |
ENST00000258411.3
|
WNT10A
|
wingless-type MMTV integration site family, member 10A |
| chr12_-_50616382 | 0.19 |
ENST00000552783.1
|
LIMA1
|
LIM domain and actin binding 1 |
| chr18_-_46987000 | 0.19 |
ENST00000442713.2
ENST00000269445.6 |
DYM
|
dymeclin |
| chr5_-_94890648 | 0.18 |
ENST00000513823.1
ENST00000514952.1 ENST00000358746.2 |
TTC37
|
tetratricopeptide repeat domain 37 |
| chr9_+_112542572 | 0.18 |
ENST00000374530.3
|
PALM2-AKAP2
|
PALM2-AKAP2 readthrough |
| chr16_+_56642489 | 0.18 |
ENST00000561491.1
|
MT2A
|
metallothionein 2A |
| chr7_+_128379346 | 0.18 |
ENST00000535011.2
ENST00000542996.2 ENST00000535623.1 ENST00000538546.1 ENST00000249364.4 ENST00000449187.2 |
CALU
|
calumenin |
| chr4_+_75310851 | 0.18 |
ENST00000395748.3
ENST00000264487.2 |
AREG
|
amphiregulin |
| chr16_+_57220049 | 0.17 |
ENST00000562439.1
|
RSPRY1
|
ring finger and SPRY domain containing 1 |
| chr10_-_105437909 | 0.17 |
ENST00000540321.1
|
SH3PXD2A
|
SH3 and PX domains 2A |
| chr14_-_35344093 | 0.17 |
ENST00000382422.2
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A |
| chr4_-_169931231 | 0.17 |
ENST00000504561.1
|
CBR4
|
carbonyl reductase 4 |
| chr1_+_222791417 | 0.17 |
ENST00000344922.5
ENST00000344441.6 ENST00000344507.1 |
MIA3
|
melanoma inhibitory activity family, member 3 |
| chrX_-_7895755 | 0.16 |
ENST00000444736.1
ENST00000537427.1 ENST00000442940.1 |
PNPLA4
|
patatin-like phospholipase domain containing 4 |
| chr1_-_27339317 | 0.16 |
ENST00000289166.5
|
FAM46B
|
family with sequence similarity 46, member B |
| chr17_+_66511540 | 0.16 |
ENST00000588188.2
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr2_-_202645612 | 0.15 |
ENST00000409632.2
ENST00000410052.1 ENST00000467448.1 |
ALS2
|
amyotrophic lateral sclerosis 2 (juvenile) |
| chr3_-_119182453 | 0.15 |
ENST00000491685.1
ENST00000461654.1 |
TMEM39A
|
transmembrane protein 39A |
| chr17_+_66511224 | 0.15 |
ENST00000588178.1
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr12_+_107349606 | 0.15 |
ENST00000547242.1
ENST00000551489.1 ENST00000550344.1 |
C12orf23
|
chromosome 12 open reading frame 23 |
| chr3_+_133292574 | 0.15 |
ENST00000264993.3
|
CDV3
|
CDV3 homolog (mouse) |
| chr16_-_11692320 | 0.14 |
ENST00000571627.1
|
LITAF
|
lipopolysaccharide-induced TNF factor |
| chr1_-_42800860 | 0.14 |
ENST00000445886.1
ENST00000361346.1 ENST00000361776.1 |
FOXJ3
|
forkhead box J3 |
| chr2_+_192542879 | 0.14 |
ENST00000409510.1
|
NABP1
|
nucleic acid binding protein 1 |
| chr12_+_1100449 | 0.14 |
ENST00000360905.4
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr12_-_76953573 | 0.14 |
ENST00000549646.1
ENST00000550628.1 ENST00000553139.1 ENST00000261183.3 ENST00000393250.4 |
OSBPL8
|
oxysterol binding protein-like 8 |
| chr13_-_52026730 | 0.14 |
ENST00000420668.2
|
INTS6
|
integrator complex subunit 6 |
| chr7_-_30066233 | 0.14 |
ENST00000222803.5
|
FKBP14
|
FK506 binding protein 14, 22 kDa |
| chr2_+_171785824 | 0.14 |
ENST00000452526.2
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa |
| chr3_-_121468513 | 0.14 |
ENST00000494517.1
ENST00000393667.3 |
GOLGB1
|
golgin B1 |
| chr12_-_76953513 | 0.13 |
ENST00000547540.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr5_-_131132658 | 0.13 |
ENST00000514667.1
ENST00000511848.1 ENST00000510461.1 |
CTC-432M15.3
FNIP1
|
Folliculin-interacting protein 1 folliculin interacting protein 1 |
| chr2_-_33824336 | 0.13 |
ENST00000431950.1
ENST00000403368.1 ENST00000441530.2 |
FAM98A
|
family with sequence similarity 98, member A |
| chr3_-_139108475 | 0.13 |
ENST00000515006.1
ENST00000513274.1 ENST00000514508.1 ENST00000507777.1 ENST00000512153.1 ENST00000333188.5 |
COPB2
|
coatomer protein complex, subunit beta 2 (beta prime) |
| chr2_-_33824382 | 0.13 |
ENST00000238823.8
|
FAM98A
|
family with sequence similarity 98, member A |
| chr12_+_1100370 | 0.13 |
ENST00000543086.3
ENST00000546231.2 ENST00000397203.2 |
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr2_+_192542850 | 0.13 |
ENST00000410026.2
|
NABP1
|
nucleic acid binding protein 1 |
| chr5_-_131562935 | 0.13 |
ENST00000379104.2
ENST00000379100.2 ENST00000428369.1 |
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chr1_+_26758849 | 0.12 |
ENST00000533087.1
ENST00000531312.1 ENST00000525165.1 ENST00000525326.1 ENST00000525546.1 ENST00000436153.2 ENST00000530781.1 |
DHDDS
|
dehydrodolichyl diphosphate synthase |
| chr12_+_1100423 | 0.12 |
ENST00000592048.1
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr11_-_118927880 | 0.12 |
ENST00000527038.2
|
HYOU1
|
hypoxia up-regulated 1 |
| chr13_+_113951607 | 0.12 |
ENST00000397181.3
|
LAMP1
|
lysosomal-associated membrane protein 1 |
| chr3_-_178789220 | 0.12 |
ENST00000414084.1
|
ZMAT3
|
zinc finger, matrin-type 3 |
| chr1_-_42801540 | 0.12 |
ENST00000372573.1
|
FOXJ3
|
forkhead box J3 |
| chr19_-_58459039 | 0.12 |
ENST00000282308.3
ENST00000598928.1 |
ZNF256
|
zinc finger protein 256 |
| chr17_+_75452447 | 0.12 |
ENST00000591472.2
|
SEPT9
|
septin 9 |
| chr8_+_64081214 | 0.12 |
ENST00000542911.2
|
YTHDF3
|
YTH domain family, member 3 |
| chr18_+_56531584 | 0.12 |
ENST00000590287.1
|
ZNF532
|
zinc finger protein 532 |
| chr13_-_36920615 | 0.12 |
ENST00000494062.2
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome) |
| chr16_+_56642041 | 0.12 |
ENST00000245185.5
|
MT2A
|
metallothionein 2A |
| chr11_-_118927735 | 0.12 |
ENST00000526656.1
|
HYOU1
|
hypoxia up-regulated 1 |
| chr3_-_139108463 | 0.11 |
ENST00000512242.1
|
COPB2
|
coatomer protein complex, subunit beta 2 (beta prime) |
| chr13_+_113951532 | 0.11 |
ENST00000332556.4
|
LAMP1
|
lysosomal-associated membrane protein 1 |
| chr18_-_47017956 | 0.11 |
ENST00000584895.1
ENST00000332968.6 ENST00000580210.1 ENST00000579408.1 |
RPL17-C18orf32
RPL17
|
RPL17-C18orf32 readthrough ribosomal protein L17 |
| chr13_-_36920420 | 0.11 |
ENST00000438666.2
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome) |
| chr2_-_242211359 | 0.11 |
ENST00000444092.1
|
HDLBP
|
high density lipoprotein binding protein |
| chr6_+_32936353 | 0.11 |
ENST00000374825.4
|
BRD2
|
bromodomain containing 2 |
| chr3_+_37903432 | 0.11 |
ENST00000443503.2
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr5_+_65440032 | 0.10 |
ENST00000334121.6
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr6_-_135375921 | 0.10 |
ENST00000367820.2
ENST00000314674.3 ENST00000524715.1 ENST00000415177.2 ENST00000367826.2 |
HBS1L
|
HBS1-like (S. cerevisiae) |
| chr12_-_76953453 | 0.10 |
ENST00000549570.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr1_-_38156153 | 0.10 |
ENST00000464085.1
ENST00000486637.1 ENST00000358011.4 ENST00000461359.1 |
C1orf109
|
chromosome 1 open reading frame 109 |
| chr10_-_120840309 | 0.10 |
ENST00000369144.3
|
EIF3A
|
eukaryotic translation initiation factor 3, subunit A |
| chr1_+_162531294 | 0.10 |
ENST00000367926.4
ENST00000271469.3 |
UAP1
|
UDP-N-acteylglucosamine pyrophosphorylase 1 |
| chr11_+_118443098 | 0.10 |
ENST00000392859.3
ENST00000359415.4 ENST00000534182.2 ENST00000264028.4 |
ARCN1
|
archain 1 |
| chr12_-_93835665 | 0.09 |
ENST00000552442.1
ENST00000550657.1 |
UBE2N
|
ubiquitin-conjugating enzyme E2N |
| chr10_+_64893039 | 0.09 |
ENST00000277746.6
ENST00000435510.2 |
NRBF2
|
nuclear receptor binding factor 2 |
| chr10_+_129785536 | 0.09 |
ENST00000419012.2
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E |
| chr11_-_3818688 | 0.09 |
ENST00000355260.3
ENST00000397004.4 ENST00000397007.4 ENST00000532475.1 |
NUP98
|
nucleoporin 98kDa |
| chr19_+_18668616 | 0.09 |
ENST00000600372.1
|
KXD1
|
KxDL motif containing 1 |
| chr6_-_90062543 | 0.09 |
ENST00000435041.2
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1 |
| chr4_+_56262115 | 0.09 |
ENST00000506198.1
ENST00000381334.5 ENST00000542052.1 |
TMEM165
|
transmembrane protein 165 |
| chr2_-_202645835 | 0.09 |
ENST00000264276.6
|
ALS2
|
amyotrophic lateral sclerosis 2 (juvenile) |
| chr5_-_131563501 | 0.09 |
ENST00000401867.1
ENST00000379086.1 ENST00000418055.1 ENST00000453286.1 ENST00000166534.4 |
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chr11_-_118927816 | 0.09 |
ENST00000534233.1
ENST00000532752.1 ENST00000525859.1 ENST00000404233.3 ENST00000532421.1 ENST00000543287.1 ENST00000527310.2 ENST00000529972.1 |
HYOU1
|
hypoxia up-regulated 1 |
| chr12_-_113909877 | 0.09 |
ENST00000261731.3
|
LHX5
|
LIM homeobox 5 |
| chr3_-_121468602 | 0.09 |
ENST00000340645.5
|
GOLGB1
|
golgin B1 |
| chr6_-_44225231 | 0.09 |
ENST00000538577.1
ENST00000537814.1 ENST00000393810.1 ENST00000393812.3 |
SLC35B2
|
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B2 |
| chr6_-_86353510 | 0.09 |
ENST00000444272.1
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr1_+_27669719 | 0.08 |
ENST00000473280.1
|
SYTL1
|
synaptotagmin-like 1 |
| chr5_+_109025067 | 0.08 |
ENST00000261483.4
|
MAN2A1
|
mannosidase, alpha, class 2A, member 1 |
| chr12_-_54121261 | 0.08 |
ENST00000549784.1
ENST00000262059.4 |
CALCOCO1
|
calcium binding and coiled-coil domain 1 |
| chr1_+_160313165 | 0.08 |
ENST00000421914.1
ENST00000535857.1 ENST00000438008.1 |
NCSTN
|
nicastrin |
| chr5_+_94890778 | 0.08 |
ENST00000380009.4
|
ARSK
|
arylsulfatase family, member K |
| chr6_+_151773583 | 0.08 |
ENST00000545879.1
|
C6orf211
|
chromosome 6 open reading frame 211 |
| chr7_+_30174574 | 0.08 |
ENST00000409688.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr13_-_36920872 | 0.08 |
ENST00000451493.1
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome) |
| chrX_-_7895479 | 0.08 |
ENST00000381042.4
|
PNPLA4
|
patatin-like phospholipase domain containing 4 |
| chr12_-_122750957 | 0.08 |
ENST00000451053.2
|
VPS33A
|
vacuolar protein sorting 33 homolog A (S. cerevisiae) |
| chr11_-_118927561 | 0.08 |
ENST00000530473.1
|
HYOU1
|
hypoxia up-regulated 1 |
| chr15_-_74988281 | 0.08 |
ENST00000566828.1
ENST00000563009.1 ENST00000568176.1 ENST00000566243.1 ENST00000566219.1 ENST00000426797.3 ENST00000566119.1 ENST00000315127.4 |
EDC3
|
enhancer of mRNA decapping 3 |
| chr11_+_69455855 | 0.08 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr6_-_101329191 | 0.08 |
ENST00000324723.6
ENST00000369162.2 ENST00000522650.1 |
ASCC3
|
activating signal cointegrator 1 complex subunit 3 |
| chr5_+_138210919 | 0.08 |
ENST00000522013.1
ENST00000520260.1 ENST00000523298.1 ENST00000520865.1 ENST00000519634.1 ENST00000517533.1 ENST00000523685.1 ENST00000519768.1 ENST00000517656.1 ENST00000521683.1 ENST00000521640.1 ENST00000519116.1 |
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr14_+_101293687 | 0.08 |
ENST00000455286.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr20_+_33292507 | 0.08 |
ENST00000414082.1
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2 |
| chr8_+_64081118 | 0.08 |
ENST00000539294.1
|
YTHDF3
|
YTH domain family, member 3 |
| chr16_-_66959429 | 0.08 |
ENST00000420652.1
ENST00000299759.6 |
RRAD
|
Ras-related associated with diabetes |
| chr4_+_128982490 | 0.08 |
ENST00000394288.3
ENST00000432347.2 ENST00000264584.5 ENST00000441387.1 ENST00000427266.1 ENST00000354456.3 |
LARP1B
|
La ribonucleoprotein domain family, member 1B |
| chr5_+_122110691 | 0.08 |
ENST00000379516.2
ENST00000505934.1 ENST00000514949.1 |
SNX2
|
sorting nexin 2 |
| chr5_-_175965008 | 0.08 |
ENST00000537487.1
|
RNF44
|
ring finger protein 44 |
| chr5_+_54603566 | 0.08 |
ENST00000230640.5
|
SKIV2L2
|
superkiller viralicidic activity 2-like 2 (S. cerevisiae) |
| chr4_+_141445333 | 0.07 |
ENST00000507667.1
|
ELMOD2
|
ELMO/CED-12 domain containing 2 |
| chr1_+_145575980 | 0.07 |
ENST00000393045.2
|
PIAS3
|
protein inhibitor of activated STAT, 3 |
| chr4_-_119757239 | 0.07 |
ENST00000280551.6
|
SEC24D
|
SEC24 family member D |
| chr5_-_150138246 | 0.07 |
ENST00000518015.1
|
DCTN4
|
dynactin 4 (p62) |
| chr4_+_128982430 | 0.07 |
ENST00000512292.1
ENST00000508819.1 |
LARP1B
|
La ribonucleoprotein domain family, member 1B |
| chr12_+_42725554 | 0.07 |
ENST00000546750.1
ENST00000547847.1 |
PPHLN1
|
periphilin 1 |
| chr1_+_228327943 | 0.07 |
ENST00000366726.1
ENST00000312726.4 ENST00000366728.2 ENST00000453943.1 ENST00000366723.1 ENST00000366722.1 ENST00000435153.1 ENST00000366721.1 |
GUK1
|
guanylate kinase 1 |
| chr20_+_57467204 | 0.07 |
ENST00000603546.1
|
GNAS
|
GNAS complex locus |
| chr22_+_21271714 | 0.07 |
ENST00000354336.3
|
CRKL
|
v-crk avian sarcoma virus CT10 oncogene homolog-like |
| chr1_+_228327923 | 0.07 |
ENST00000391865.3
|
GUK1
|
guanylate kinase 1 |
| chr13_-_31040060 | 0.07 |
ENST00000326004.4
ENST00000341423.5 |
HMGB1
|
high mobility group box 1 |
| chr15_+_84904525 | 0.07 |
ENST00000510439.2
|
GOLGA6L4
|
golgin A6 family-like 4 |
| chr10_+_129785574 | 0.07 |
ENST00000430713.2
ENST00000471218.1 |
PTPRE
|
protein tyrosine phosphatase, receptor type, E |
| chrX_+_101854426 | 0.07 |
ENST00000536530.1
ENST00000473968.1 ENST00000604957.1 ENST00000477663.1 ENST00000479502.1 |
ARMCX5
|
armadillo repeat containing, X-linked 5 |
| chr1_-_47184723 | 0.07 |
ENST00000371933.3
|
EFCAB14
|
EF-hand calcium binding domain 14 |
| chr3_-_113465065 | 0.07 |
ENST00000497255.1
ENST00000478020.1 ENST00000240922.3 ENST00000493900.1 |
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr3_+_113465866 | 0.07 |
ENST00000273398.3
ENST00000538620.1 ENST00000496747.1 ENST00000475322.1 |
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A |
| chr12_-_106641728 | 0.07 |
ENST00000378026.4
|
CKAP4
|
cytoskeleton-associated protein 4 |
| chr1_+_26758790 | 0.07 |
ENST00000427245.2
ENST00000525682.2 ENST00000236342.7 ENST00000526219.1 ENST00000374185.3 ENST00000360009.2 |
DHDDS
|
dehydrodolichyl diphosphate synthase |
| chr1_+_60280458 | 0.07 |
ENST00000455990.1
ENST00000371208.3 |
HOOK1
|
hook microtubule-tethering protein 1 |
| chr7_+_39663061 | 0.07 |
ENST00000005257.2
|
RALA
|
v-ral simian leukemia viral oncogene homolog A (ras related) |
| chr16_+_57220193 | 0.07 |
ENST00000564435.1
ENST00000562959.1 ENST00000394420.4 ENST00000568505.2 ENST00000537866.1 |
RSPRY1
|
ring finger and SPRY domain containing 1 |
| chr5_+_126853301 | 0.07 |
ENST00000296666.8
ENST00000442138.2 ENST00000512635.2 |
PRRC1
|
proline-rich coiled-coil 1 |
| chr19_+_2841433 | 0.07 |
ENST00000334241.4
ENST00000585966.1 ENST00000591539.1 |
ZNF555
|
zinc finger protein 555 |
| chr6_+_44355257 | 0.06 |
ENST00000371477.3
|
CDC5L
|
cell division cycle 5-like |
| chr1_+_44445643 | 0.06 |
ENST00000309519.7
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr3_-_4508925 | 0.06 |
ENST00000534863.1
ENST00000383843.5 ENST00000458465.2 ENST00000405420.2 ENST00000272902.5 |
SUMF1
|
sulfatase modifying factor 1 |
| chr3_+_128598433 | 0.06 |
ENST00000308982.7
ENST00000514336.1 |
ACAD9
|
acyl-CoA dehydrogenase family, member 9 |
| chr3_-_57583130 | 0.06 |
ENST00000303436.6
|
ARF4
|
ADP-ribosylation factor 4 |
| chr6_-_43337180 | 0.06 |
ENST00000318149.3
ENST00000361428.2 |
ZNF318
|
zinc finger protein 318 |
| chr7_+_30174426 | 0.06 |
ENST00000324453.8
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chrX_-_46618490 | 0.06 |
ENST00000328306.4
|
SLC9A7
|
solute carrier family 9, subfamily A (NHE7, cation proton antiporter 7), member 7 |
| chr11_-_119234876 | 0.06 |
ENST00000525735.1
|
USP2
|
ubiquitin specific peptidase 2 |
| chr18_-_46895066 | 0.06 |
ENST00000583225.1
ENST00000584983.1 ENST00000583280.1 ENST00000581738.1 |
DYM
|
dymeclin |
| chr22_-_28392227 | 0.06 |
ENST00000431039.1
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr17_-_6554877 | 0.06 |
ENST00000225728.3
ENST00000575197.1 |
MED31
|
mediator complex subunit 31 |
| chr7_+_30174668 | 0.06 |
ENST00000415604.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr19_+_16187816 | 0.06 |
ENST00000588410.1
|
TPM4
|
tropomyosin 4 |
| chr1_+_160313062 | 0.06 |
ENST00000294785.5
ENST00000368063.1 ENST00000437169.1 |
NCSTN
|
nicastrin |
| chr17_-_6554747 | 0.06 |
ENST00000574128.1
|
MED31
|
mediator complex subunit 31 |
| chr10_-_71930222 | 0.06 |
ENST00000458634.2
ENST00000373239.2 ENST00000373242.2 ENST00000373241.4 |
SAR1A
|
SAR1 homolog A (S. cerevisiae) |
| chr1_+_249200395 | 0.06 |
ENST00000355360.4
ENST00000329291.5 ENST00000539153.1 |
PGBD2
|
piggyBac transposable element derived 2 |
| chr19_+_10527449 | 0.05 |
ENST00000592685.1
ENST00000380702.2 |
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chrX_+_134478706 | 0.05 |
ENST00000370761.3
ENST00000339249.4 ENST00000370760.3 |
ZNF449
|
zinc finger protein 449 |
| chr3_-_107941209 | 0.05 |
ENST00000492106.1
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr3_+_134204881 | 0.05 |
ENST00000511574.1
ENST00000337090.3 ENST00000383229.3 |
CEP63
|
centrosomal protein 63kDa |
| chr12_-_53473136 | 0.05 |
ENST00000547837.1
ENST00000301463.4 |
SPRYD3
|
SPRY domain containing 3 |
| chr5_-_114961673 | 0.05 |
ENST00000333314.3
|
TMED7-TICAM2
|
TMED7-TICAM2 readthrough |
| chr3_-_33759541 | 0.05 |
ENST00000468888.2
|
CLASP2
|
cytoplasmic linker associated protein 2 |
| chrX_+_134124968 | 0.05 |
ENST00000330288.4
|
SMIM10
|
small integral membrane protein 10 |
| chr5_-_150138551 | 0.05 |
ENST00000446090.2
ENST00000447998.2 |
DCTN4
|
dynactin 4 (p62) |
| chr1_+_145576007 | 0.05 |
ENST00000369298.1
|
PIAS3
|
protein inhibitor of activated STAT, 3 |
| chr2_-_69614373 | 0.05 |
ENST00000361060.5
ENST00000357308.4 |
GFPT1
|
glutamine--fructose-6-phosphate transaminase 1 |
| chr14_+_68086515 | 0.05 |
ENST00000261783.3
|
ARG2
|
arginase 2 |
| chr16_-_69390209 | 0.05 |
ENST00000563634.1
|
RP11-343C2.9
|
Uncharacterized protein |
| chr12_+_1100710 | 0.05 |
ENST00000589132.1
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr3_-_47823298 | 0.05 |
ENST00000254480.5
|
SMARCC1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 |
| chr20_+_55926566 | 0.05 |
ENST00000411894.1
ENST00000429339.1 |
RAE1
|
ribonucleic acid export 1 |
| chr10_+_121410882 | 0.05 |
ENST00000369085.3
|
BAG3
|
BCL2-associated athanogene 3 |
| chr12_+_44229846 | 0.05 |
ENST00000551577.1
ENST00000266534.3 |
TMEM117
|
transmembrane protein 117 |
| chr3_-_57583185 | 0.05 |
ENST00000463880.1
|
ARF4
|
ADP-ribosylation factor 4 |
| chr18_+_76829441 | 0.05 |
ENST00000458297.2
|
ATP9B
|
ATPase, class II, type 9B |
| chr3_+_19988885 | 0.05 |
ENST00000422242.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr3_-_33759699 | 0.05 |
ENST00000399362.4
ENST00000359576.5 ENST00000307312.7 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr2_-_198299726 | 0.05 |
ENST00000409915.4
ENST00000487698.1 ENST00000414963.2 ENST00000335508.6 |
SF3B1
|
splicing factor 3b, subunit 1, 155kDa |
| chr22_+_50981079 | 0.05 |
ENST00000609268.1
|
CTA-384D8.34
|
CTA-384D8.34 |
| chr2_+_17997763 | 0.04 |
ENST00000281047.3
|
MSGN1
|
mesogenin 1 |
| chr10_-_111683308 | 0.04 |
ENST00000502935.1
ENST00000322238.8 ENST00000369680.4 |
XPNPEP1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chr9_+_117350009 | 0.04 |
ENST00000374050.3
|
ATP6V1G1
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G1 |
| chr1_+_44445549 | 0.04 |
ENST00000356836.6
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr3_+_134205000 | 0.04 |
ENST00000512894.1
ENST00000513612.2 ENST00000606977.1 |
CEP63
|
centrosomal protein 63kDa |
| chr3_+_47422485 | 0.04 |
ENST00000431726.1
ENST00000456221.1 ENST00000265562.4 |
PTPN23
|
protein tyrosine phosphatase, non-receptor type 23 |
| chr15_+_76135622 | 0.04 |
ENST00000338677.4
ENST00000267938.4 ENST00000569423.1 |
UBE2Q2
|
ubiquitin-conjugating enzyme E2Q family member 2 |
| chr5_-_54603368 | 0.04 |
ENST00000508346.1
ENST00000251636.5 |
DHX29
|
DEAH (Asp-Glu-Ala-His) box polypeptide 29 |
| chr13_-_45915221 | 0.04 |
ENST00000309246.5
ENST00000379060.4 ENST00000379055.1 ENST00000527226.1 ENST00000379056.1 |
TPT1
|
tumor protein, translationally-controlled 1 |
| chr4_+_128982416 | 0.04 |
ENST00000326639.6
|
LARP1B
|
La ribonucleoprotein domain family, member 1B |
| chr18_-_47018897 | 0.04 |
ENST00000418495.1
|
RPL17
|
ribosomal protein L17 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CREB3L1_CREB3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0015728 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.1 | 0.4 | GO:0036483 | neuron intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0036483) regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903381) negative regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903382) |
| 0.1 | 0.2 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 | 0.3 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 | 0.3 | GO:0010979 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.1 | 0.3 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.1 | 0.3 | GO:0043553 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.2 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.0 | 0.2 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.4 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.3 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.4 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.1 | GO:0046963 | 3'-phosphoadenosine 5'-phosphosulfate transport(GO:0046963) 3'-phospho-5'-adenylyl sulfate transmembrane transport(GO:1902559) |
| 0.0 | 0.1 | GO:0009183 | purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 0.0 | 0.6 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.3 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.7 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.2 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.2 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.0 | 0.4 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.0 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.4 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.1 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) |
| 0.0 | 0.3 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 0.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.1 | GO:0030011 | maintenance of cell polarity(GO:0030011) maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.2 | GO:0030638 | polyketide metabolic process(GO:0030638) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.2 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.4 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.0 | 0.0 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.1 | 0.2 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 0.2 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.0 | GO:0070993 | translation preinitiation complex(GO:0070993) |
| 0.0 | 0.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.5 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.0 | 0.2 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.2 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.1 | 0.7 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 0.3 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.1 | 0.2 | GO:0003955 | NAD(P)H dehydrogenase (quinone) activity(GO:0003955) 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.2 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.2 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.4 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.3 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0000224 | peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase activity(GO:0000224) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.3 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.1 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.0 | 0.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |