Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
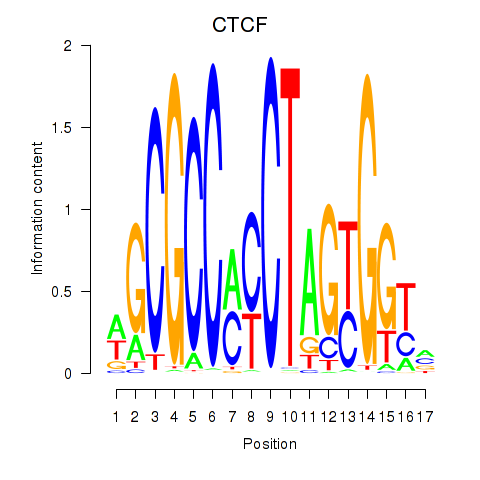
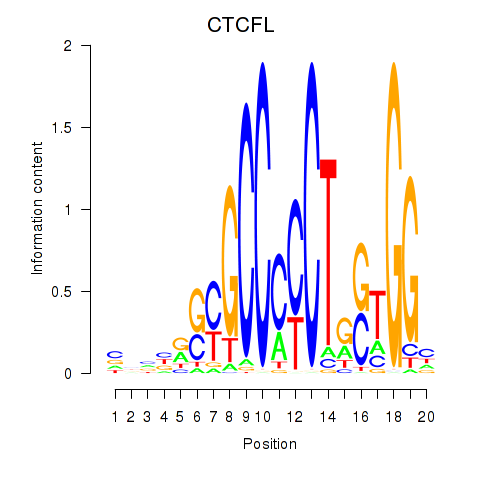
Results for CTCF_CTCFL
Z-value: 2.40
Transcription factors associated with CTCF_CTCFL
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CTCF
|
ENSG00000102974.10 | CCCTC-binding factor |
|
CTCFL
|
ENSG00000124092.8 | CCCTC-binding factor like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CTCFL | hg19_v2_chr20_-_56100179_56100265 | 0.16 | 5.1e-01 | Click! |
| CTCF | hg19_v2_chr16_+_67596310_67596353 | 0.10 | 6.8e-01 | Click! |
Activity profile of CTCF_CTCFL motif
Sorted Z-values of CTCF_CTCFL motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_2390704 | 15.81 |
ENST00000301732.5
ENST00000382381.3 |
ABCA3
|
ATP-binding cassette, sub-family A (ABC1), member 3 |
| chr22_-_22901636 | 14.87 |
ENST00000406503.1
ENST00000439106.1 ENST00000402697.1 ENST00000543184.1 ENST00000398743.2 |
PRAME
|
preferentially expressed antigen in melanoma |
| chr22_-_22901477 | 13.66 |
ENST00000420709.1
ENST00000398741.1 ENST00000405655.3 |
PRAME
|
preferentially expressed antigen in melanoma |
| chr11_+_61520075 | 13.33 |
ENST00000278836.5
|
MYRF
|
myelin regulatory factor |
| chr9_+_130965651 | 12.27 |
ENST00000475805.1
ENST00000341179.7 ENST00000372923.3 |
DNM1
|
dynamin 1 |
| chr5_+_175299743 | 11.96 |
ENST00000502265.1
|
CPLX2
|
complexin 2 |
| chr9_+_130965677 | 11.04 |
ENST00000393594.3
ENST00000486160.1 |
DNM1
|
dynamin 1 |
| chr11_-_61584233 | 10.36 |
ENST00000491310.1
|
FADS1
|
fatty acid desaturase 1 |
| chr5_+_176513895 | 9.00 |
ENST00000503708.1
ENST00000393648.2 ENST00000514472.1 ENST00000502906.1 ENST00000292410.3 ENST00000510911.1 |
FGFR4
|
fibroblast growth factor receptor 4 |
| chrX_-_30326445 | 8.92 |
ENST00000378963.1
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr20_+_48807351 | 8.87 |
ENST00000303004.3
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta |
| chr17_+_74864476 | 8.78 |
ENST00000301618.4
ENST00000569840.2 |
MGAT5B
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B |
| chr2_+_130939235 | 8.37 |
ENST00000425361.1
ENST00000457492.1 |
MZT2B
|
mitotic spindle organizing protein 2B |
| chr2_+_27309605 | 8.05 |
ENST00000260599.6
ENST00000260598.5 ENST00000429697.1 |
KHK
|
ketohexokinase (fructokinase) |
| chr16_+_30996502 | 7.68 |
ENST00000353250.5
ENST00000262520.6 ENST00000297679.5 ENST00000562932.1 ENST00000574447.1 |
HSD3B7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr2_-_132249955 | 7.34 |
ENST00000309451.6
|
MZT2A
|
mitotic spindle organizing protein 2A |
| chr2_+_130939827 | 7.32 |
ENST00000409255.1
ENST00000455239.1 |
MZT2B
|
mitotic spindle organizing protein 2B |
| chr8_-_48651648 | 7.24 |
ENST00000408965.3
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr19_+_507299 | 7.21 |
ENST00000359315.5
|
TPGS1
|
tubulin polyglutamylase complex subunit 1 |
| chr8_+_17434689 | 7.04 |
ENST00000398074.3
|
PDGFRL
|
platelet-derived growth factor receptor-like |
| chr20_-_34542548 | 6.95 |
ENST00000305978.2
|
SCAND1
|
SCAN domain containing 1 |
| chr5_+_176513868 | 6.94 |
ENST00000292408.4
|
FGFR4
|
fibroblast growth factor receptor 4 |
| chr17_+_43213004 | 6.70 |
ENST00000586346.1
ENST00000398322.3 ENST00000592162.1 ENST00000376955.4 ENST00000321854.8 |
ACBD4
|
acyl-CoA binding domain containing 4 |
| chr9_+_131464767 | 6.55 |
ENST00000291906.4
|
PKN3
|
protein kinase N3 |
| chr16_+_1203194 | 6.29 |
ENST00000348261.5
ENST00000358590.4 |
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit |
| chr16_+_67313412 | 6.18 |
ENST00000379344.3
ENST00000568621.1 ENST00000450733.1 ENST00000567938.1 |
PLEKHG4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| chr19_+_38893809 | 5.88 |
ENST00000589408.1
|
FAM98C
|
family with sequence similarity 98, member C |
| chr12_-_7245125 | 5.77 |
ENST00000542285.1
ENST00000540610.1 |
C1R
|
complement component 1, r subcomponent |
| chr14_+_31343747 | 5.68 |
ENST00000216361.4
ENST00000396618.3 ENST00000475087.1 |
COCH
|
cochlin |
| chr16_+_230435 | 5.57 |
ENST00000199708.2
|
HBQ1
|
hemoglobin, theta 1 |
| chr16_+_71392616 | 5.49 |
ENST00000349553.5
ENST00000302628.4 ENST00000562305.1 |
CALB2
|
calbindin 2 |
| chr7_+_302918 | 5.46 |
ENST00000599994.1
|
AC187652.1
|
Protein LOC100996433 |
| chr20_-_33460621 | 5.45 |
ENST00000427420.1
ENST00000336431.5 |
GGT7
|
gamma-glutamyltransferase 7 |
| chr9_-_35096545 | 5.43 |
ENST00000378617.3
ENST00000341666.3 ENST00000361778.2 |
PIGO
|
phosphatidylinositol glycan anchor biosynthesis, class O |
| chr19_-_40030861 | 5.32 |
ENST00000390658.2
|
EID2
|
EP300 interacting inhibitor of differentiation 2 |
| chr11_+_64781575 | 5.04 |
ENST00000246747.4
ENST00000529384.1 |
ARL2
|
ADP-ribosylation factor-like 2 |
| chr1_+_212208919 | 5.04 |
ENST00000366991.4
ENST00000542077.1 |
DTL
|
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr5_+_140254884 | 5.02 |
ENST00000398631.2
|
PCDHA12
|
protocadherin alpha 12 |
| chr9_+_140317802 | 4.96 |
ENST00000341349.2
ENST00000392815.2 |
NOXA1
|
NADPH oxidase activator 1 |
| chr12_-_7245018 | 4.95 |
ENST00000543835.1
ENST00000535233.2 |
C1R
|
complement component 1, r subcomponent |
| chr16_+_84002234 | 4.92 |
ENST00000305202.4
|
NECAB2
|
N-terminal EF-hand calcium binding protein 2 |
| chr1_+_64239657 | 4.88 |
ENST00000371080.1
ENST00000371079.1 |
ROR1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr19_-_38747172 | 4.87 |
ENST00000347262.4
ENST00000591585.1 ENST00000301242.4 |
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr17_+_7591747 | 4.86 |
ENST00000534050.1
|
WRAP53
|
WD repeat containing, antisense to TP53 |
| chr17_-_19266045 | 4.77 |
ENST00000395616.3
|
B9D1
|
B9 protein domain 1 |
| chr5_+_140186647 | 4.76 |
ENST00000512229.2
ENST00000356878.4 ENST00000530339.1 |
PCDHA4
|
protocadherin alpha 4 |
| chr12_-_7245080 | 4.75 |
ENST00000541042.1
ENST00000540242.1 |
C1R
|
complement component 1, r subcomponent |
| chr17_+_48556158 | 4.69 |
ENST00000258955.2
|
RSAD1
|
radical S-adenosyl methionine domain containing 1 |
| chr20_+_3767547 | 4.69 |
ENST00000344256.6
ENST00000379598.5 |
CDC25B
|
cell division cycle 25B |
| chr19_+_38893751 | 4.62 |
ENST00000588262.1
ENST00000252530.5 ENST00000343358.7 |
FAM98C
|
family with sequence similarity 98, member C |
| chr12_-_7245152 | 4.62 |
ENST00000542220.2
|
C1R
|
complement component 1, r subcomponent |
| chr12_-_58027138 | 4.56 |
ENST00000341156.4
|
B4GALNT1
|
beta-1,4-N-acetyl-galactosaminyl transferase 1 |
| chr1_+_203830703 | 4.55 |
ENST00000414487.2
|
SNRPE
|
small nuclear ribonucleoprotein polypeptide E |
| chr22_-_30956746 | 4.53 |
ENST00000437282.1
ENST00000447224.1 ENST00000427899.1 ENST00000406955.1 ENST00000452827.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr8_-_90996837 | 4.49 |
ENST00000519426.1
ENST00000265433.3 |
NBN
|
nibrin |
| chr7_+_73082152 | 4.48 |
ENST00000324941.4
ENST00000451519.1 |
VPS37D
|
vacuolar protein sorting 37 homolog D (S. cerevisiae) |
| chr17_+_7591639 | 4.38 |
ENST00000396463.2
|
WRAP53
|
WD repeat containing, antisense to TP53 |
| chr2_+_20866424 | 4.35 |
ENST00000272224.3
|
GDF7
|
growth differentiation factor 7 |
| chr7_-_27219849 | 4.34 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr17_+_16593539 | 4.29 |
ENST00000340621.5
ENST00000399273.1 ENST00000443444.2 ENST00000360524.8 ENST00000456009.1 |
CCDC144A
|
coiled-coil domain containing 144A |
| chr15_+_99645277 | 4.28 |
ENST00000336292.6
ENST00000328642.7 |
SYNM
|
synemin, intermediate filament protein |
| chr19_-_38746979 | 4.24 |
ENST00000591291.1
|
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr17_-_42441204 | 4.20 |
ENST00000293443.7
|
FAM171A2
|
family with sequence similarity 171, member A2 |
| chr7_-_16840820 | 4.16 |
ENST00000450569.1
|
AGR2
|
anterior gradient 2 |
| chr15_-_34502278 | 4.15 |
ENST00000559515.1
ENST00000256544.3 ENST00000560108.1 ENST00000559462.1 |
KATNBL1
|
katanin p80 subunit B-like 1 |
| chr6_+_33359582 | 4.14 |
ENST00000450504.1
|
KIFC1
|
kinesin family member C1 |
| chr20_+_48599506 | 4.08 |
ENST00000244050.2
|
SNAI1
|
snail family zinc finger 1 |
| chr19_+_18699535 | 4.04 |
ENST00000358607.6
|
C19orf60
|
chromosome 19 open reading frame 60 |
| chr17_+_78075498 | 4.03 |
ENST00000302262.3
|
GAA
|
glucosidase, alpha; acid |
| chr6_+_32821924 | 4.01 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr4_-_141348999 | 3.99 |
ENST00000325617.5
|
CLGN
|
calmegin |
| chr12_-_58026920 | 3.99 |
ENST00000550764.1
ENST00000551220.1 |
B4GALNT1
|
beta-1,4-N-acetyl-galactosaminyl transferase 1 |
| chr7_+_89783689 | 3.93 |
ENST00000297205.2
|
STEAP1
|
six transmembrane epithelial antigen of the prostate 1 |
| chr16_+_19179549 | 3.91 |
ENST00000355377.2
ENST00000568115.1 |
SYT17
|
synaptotagmin XVII |
| chr20_-_25038804 | 3.90 |
ENST00000323482.4
|
ACSS1
|
acyl-CoA synthetase short-chain family member 1 |
| chr15_-_64338521 | 3.87 |
ENST00000457488.1
ENST00000558069.1 |
DAPK2
|
death-associated protein kinase 2 |
| chr22_-_31503490 | 3.78 |
ENST00000400299.2
|
SELM
|
Selenoprotein M |
| chr8_-_90996459 | 3.70 |
ENST00000517337.1
ENST00000409330.1 |
NBN
|
nibrin |
| chr2_+_234263120 | 3.66 |
ENST00000264057.2
ENST00000427930.1 |
DGKD
|
diacylglycerol kinase, delta 130kDa |
| chr11_+_61447845 | 3.64 |
ENST00000257215.5
|
DAGLA
|
diacylglycerol lipase, alpha |
| chr14_+_105957402 | 3.64 |
ENST00000421892.1
ENST00000334656.7 ENST00000451719.1 ENST00000392522.3 ENST00000392523.4 ENST00000354560.6 ENST00000450383.1 |
C14orf80
|
chromosome 14 open reading frame 80 |
| chr16_-_19897455 | 3.63 |
ENST00000568214.1
ENST00000569479.1 |
GPRC5B
|
G protein-coupled receptor, family C, group 5, member B |
| chr17_-_19265982 | 3.63 |
ENST00000268841.6
ENST00000261499.4 ENST00000575478.1 |
B9D1
|
B9 protein domain 1 |
| chr8_-_27850180 | 3.56 |
ENST00000380385.2
ENST00000301906.4 ENST00000354914.3 |
SCARA5
|
scavenger receptor class A, member 5 (putative) |
| chr4_-_141348789 | 3.54 |
ENST00000414773.1
|
CLGN
|
calmegin |
| chr12_+_48357340 | 3.53 |
ENST00000256686.6
ENST00000549288.1 ENST00000552561.1 ENST00000546749.1 ENST00000552546.1 ENST00000550552.1 |
TMEM106C
|
transmembrane protein 106C |
| chr10_+_6622345 | 3.53 |
ENST00000445427.1
ENST00000455810.1 |
PRKCQ-AS1
|
PRKCQ antisense RNA 1 |
| chr1_+_10490127 | 3.52 |
ENST00000602787.1
ENST00000602296.1 ENST00000400900.2 |
APITD1
APITD1-CORT
|
apoptosis-inducing, TAF9-like domain 1 APITD1-CORT readthrough |
| chr11_+_64781657 | 3.48 |
ENST00000533729.1
|
ARL2
|
ADP-ribosylation factor-like 2 |
| chr11_-_8285405 | 3.45 |
ENST00000335790.3
ENST00000534484.1 |
LMO1
|
LIM domain only 1 (rhombotin 1) |
| chr11_+_61583968 | 3.45 |
ENST00000517839.1
|
FADS2
|
fatty acid desaturase 2 |
| chr11_-_65150103 | 3.43 |
ENST00000294187.6
ENST00000398802.1 ENST00000360662.3 ENST00000377152.2 ENST00000530936.1 |
SLC25A45
|
solute carrier family 25, member 45 |
| chr2_+_69240302 | 3.43 |
ENST00000303714.4
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr12_-_58027002 | 3.42 |
ENST00000449184.3
|
B4GALNT1
|
beta-1,4-N-acetyl-galactosaminyl transferase 1 |
| chr8_-_145911188 | 3.42 |
ENST00000540274.1
|
ARHGAP39
|
Rho GTPase activating protein 39 |
| chr4_+_154265784 | 3.42 |
ENST00000240488.3
|
MND1
|
meiotic nuclear divisions 1 homolog (S. cerevisiae) |
| chr8_+_41386725 | 3.38 |
ENST00000276533.3
ENST00000520710.1 ENST00000518671.1 |
GINS4
|
GINS complex subunit 4 (Sld5 homolog) |
| chr19_-_55652290 | 3.28 |
ENST00000589745.1
|
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr19_+_5823813 | 3.26 |
ENST00000303212.2
|
NRTN
|
neurturin |
| chr17_+_63096903 | 3.22 |
ENST00000582940.1
|
RP11-160O5.1
|
RP11-160O5.1 |
| chr16_-_54963026 | 3.20 |
ENST00000560208.1
ENST00000557792.1 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr22_-_30783075 | 3.20 |
ENST00000215798.6
|
RNF215
|
ring finger protein 215 |
| chr19_-_39421377 | 3.19 |
ENST00000430193.3
ENST00000600042.1 ENST00000221431.6 |
SARS2
|
seryl-tRNA synthetase 2, mitochondrial |
| chr12_+_58120044 | 3.18 |
ENST00000542466.2
|
AGAP2-AS1
|
AGAP2 antisense RNA 1 |
| chr17_+_78075361 | 3.18 |
ENST00000577106.1
ENST00000390015.3 |
GAA
|
glucosidase, alpha; acid |
| chr2_+_69240511 | 3.18 |
ENST00000409349.3
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr17_-_33469299 | 3.16 |
ENST00000586869.1
ENST00000360831.5 ENST00000442241.4 |
NLE1
|
notchless homolog 1 (Drosophila) |
| chr12_+_48357401 | 3.14 |
ENST00000429772.2
ENST00000449758.2 |
TMEM106C
|
transmembrane protein 106C |
| chr9_-_130742792 | 3.13 |
ENST00000373095.1
|
FAM102A
|
family with sequence similarity 102, member A |
| chr17_-_73511584 | 3.09 |
ENST00000321617.3
|
CASKIN2
|
CASK interacting protein 2 |
| chr19_+_42363917 | 3.08 |
ENST00000598742.1
|
RPS19
|
ribosomal protein S19 |
| chr6_-_11044509 | 3.04 |
ENST00000354666.3
|
ELOVL2
|
ELOVL fatty acid elongase 2 |
| chr3_+_172468749 | 3.03 |
ENST00000366254.2
ENST00000415665.1 ENST00000438041.1 |
ECT2
|
epithelial cell transforming sequence 2 oncogene |
| chr16_-_66907139 | 3.01 |
ENST00000561579.2
|
NAE1
|
NEDD8 activating enzyme E1 subunit 1 |
| chr1_+_840205 | 3.00 |
ENST00000607769.1
|
RP11-54O7.16
|
RP11-54O7.16 |
| chr1_+_10490441 | 2.99 |
ENST00000470413.2
ENST00000309048.3 |
APITD1-CORT
APITD1
|
APITD1-CORT readthrough apoptosis-inducing, TAF9-like domain 1 |
| chr12_+_104609550 | 2.97 |
ENST00000525566.1
ENST00000429002.2 |
TXNRD1
|
thioredoxin reductase 1 |
| chr7_+_130126012 | 2.96 |
ENST00000341441.5
|
MEST
|
mesoderm specific transcript |
| chr17_+_78075324 | 2.96 |
ENST00000570803.1
|
GAA
|
glucosidase, alpha; acid |
| chr19_-_10024496 | 2.95 |
ENST00000593091.1
|
OLFM2
|
olfactomedin 2 |
| chr3_-_186857267 | 2.94 |
ENST00000455270.1
ENST00000296277.4 |
RPL39L
|
ribosomal protein L39-like |
| chr12_-_15374343 | 2.93 |
ENST00000256953.2
ENST00000546331.1 |
RERG
|
RAS-like, estrogen-regulated, growth inhibitor |
| chr11_-_18127566 | 2.91 |
ENST00000532452.1
ENST00000530180.1 ENST00000300013.4 ENST00000529318.1 ENST00000524803.1 |
SAAL1
|
serum amyloid A-like 1 |
| chr9_+_131901710 | 2.90 |
ENST00000524946.2
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr10_+_101089107 | 2.89 |
ENST00000446890.1
ENST00000370528.3 |
CNNM1
|
cyclin M1 |
| chr16_+_29827285 | 2.89 |
ENST00000320330.6
|
PAGR1
|
PAXIP1 associated glutamate-rich protein 1 |
| chr17_+_38599693 | 2.87 |
ENST00000542955.1
ENST00000269593.4 |
IGFBP4
|
insulin-like growth factor binding protein 4 |
| chr16_+_29827832 | 2.83 |
ENST00000609618.1
|
PAGR1
|
PAXIP1-associated glutamate-rich protein 1 |
| chr14_-_54908043 | 2.82 |
ENST00000556113.1
ENST00000553660.1 ENST00000395573.4 ENST00000557690.1 ENST00000216416.4 |
CNIH1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chrX_-_99891796 | 2.82 |
ENST00000373020.4
|
TSPAN6
|
tetraspanin 6 |
| chr17_-_19265855 | 2.81 |
ENST00000440841.1
ENST00000395615.1 ENST00000461069.2 |
B9D1
|
B9 protein domain 1 |
| chr2_+_69240415 | 2.80 |
ENST00000409829.3
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr19_+_46367518 | 2.79 |
ENST00000302177.2
|
FOXA3
|
forkhead box A3 |
| chr17_-_76183111 | 2.79 |
ENST00000405273.1
ENST00000590862.1 ENST00000590430.1 ENST00000586613.1 |
TK1
|
thymidine kinase 1, soluble |
| chr19_+_18699599 | 2.72 |
ENST00000450195.2
|
C19orf60
|
chromosome 19 open reading frame 60 |
| chr11_-_65149422 | 2.72 |
ENST00000526432.1
ENST00000527174.1 |
SLC25A45
|
solute carrier family 25, member 45 |
| chr19_-_3547305 | 2.69 |
ENST00000589063.1
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr22_+_25003626 | 2.68 |
ENST00000451366.1
ENST00000406383.2 ENST00000428855.1 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr4_+_2043689 | 2.65 |
ENST00000382878.3
ENST00000409248.4 |
C4orf48
|
chromosome 4 open reading frame 48 |
| chr14_+_31343951 | 2.62 |
ENST00000556908.1
ENST00000555881.1 ENST00000460581.2 |
COCH
|
cochlin |
| chr9_+_139560197 | 2.61 |
ENST00000371698.3
|
EGFL7
|
EGF-like-domain, multiple 7 |
| chr8_+_38614754 | 2.59 |
ENST00000521642.1
|
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr5_+_176514413 | 2.59 |
ENST00000513166.1
|
FGFR4
|
fibroblast growth factor receptor 4 |
| chr9_-_139258235 | 2.58 |
ENST00000371738.3
|
DNLZ
|
DNL-type zinc finger |
| chr10_+_12391685 | 2.57 |
ENST00000378845.1
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID |
| chr19_+_12949251 | 2.55 |
ENST00000251472.4
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chr9_+_131901661 | 2.54 |
ENST00000423100.1
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr17_-_33446820 | 2.54 |
ENST00000592577.1
ENST00000590016.1 ENST00000345365.6 ENST00000360276.3 ENST00000357906.3 |
RAD51D
|
RAD51 paralog D |
| chr7_+_130126165 | 2.53 |
ENST00000427521.1
ENST00000416162.2 ENST00000378576.4 |
MEST
|
mesoderm specific transcript |
| chr16_-_54962704 | 2.53 |
ENST00000502066.2
ENST00000560912.1 ENST00000558952.1 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr5_+_35617940 | 2.51 |
ENST00000282469.6
ENST00000509059.1 ENST00000356031.3 ENST00000510777.1 |
SPEF2
|
sperm flagellar 2 |
| chr12_+_53440753 | 2.51 |
ENST00000379902.3
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr3_+_48507210 | 2.50 |
ENST00000433541.1
ENST00000422277.2 ENST00000436480.2 ENST00000444177.1 |
TREX1
|
three prime repair exonuclease 1 |
| chr5_+_140201183 | 2.49 |
ENST00000529619.1
ENST00000529859.1 ENST00000378126.3 |
PCDHA5
|
protocadherin alpha 5 |
| chr20_+_3801162 | 2.49 |
ENST00000379573.2
ENST00000379567.2 ENST00000455742.1 ENST00000246041.2 |
AP5S1
|
adaptor-related protein complex 5, sigma 1 subunit |
| chr8_+_38614807 | 2.48 |
ENST00000330691.6
ENST00000348567.4 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr14_-_24685246 | 2.48 |
ENST00000396833.2
ENST00000288087.7 |
MDP1
|
magnesium-dependent phosphatase 1 |
| chr4_+_4861385 | 2.46 |
ENST00000382723.4
|
MSX1
|
msh homeobox 1 |
| chr1_+_10490779 | 2.46 |
ENST00000477755.1
|
APITD1
|
apoptosis-inducing, TAF9-like domain 1 |
| chr9_+_100174232 | 2.45 |
ENST00000355295.4
|
TDRD7
|
tudor domain containing 7 |
| chr5_+_159343688 | 2.44 |
ENST00000306675.3
|
ADRA1B
|
adrenoceptor alpha 1B |
| chr3_-_118864893 | 2.44 |
ENST00000354673.2
ENST00000425327.2 |
IGSF11
|
immunoglobulin superfamily, member 11 |
| chr11_+_2421718 | 2.44 |
ENST00000380996.5
ENST00000333256.6 ENST00000380992.1 ENST00000437110.1 ENST00000435795.1 |
TSSC4
|
tumor suppressing subtransferable candidate 4 |
| chr20_+_44441271 | 2.41 |
ENST00000335046.3
ENST00000243893.6 |
UBE2C
|
ubiquitin-conjugating enzyme E2C |
| chr4_+_89378261 | 2.40 |
ENST00000264350.3
|
HERC5
|
HECT and RLD domain containing E3 ubiquitin protein ligase 5 |
| chr4_-_141348763 | 2.40 |
ENST00000509477.1
|
CLGN
|
calmegin |
| chr10_+_101088836 | 2.39 |
ENST00000356713.4
|
CNNM1
|
cyclin M1 |
| chr2_+_26395939 | 2.39 |
ENST00000401533.2
|
GAREML
|
GRB2 associated, regulator of MAPK1-like |
| chrX_+_77166172 | 2.38 |
ENST00000343533.5
ENST00000350425.4 ENST00000341514.6 |
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr12_+_58176525 | 2.38 |
ENST00000543727.1
ENST00000540550.1 ENST00000323833.8 ENST00000350762.5 ENST00000550559.1 ENST00000548851.1 ENST00000434359.1 ENST00000457189.1 |
TSFM
|
Ts translation elongation factor, mitochondrial |
| chr17_-_73901494 | 2.36 |
ENST00000309352.3
|
MRPL38
|
mitochondrial ribosomal protein L38 |
| chr12_-_109531264 | 2.36 |
ENST00000429722.2
ENST00000536242.1 ENST00000343075.3 ENST00000536358.1 |
ALKBH2
|
alkB, alkylation repair homolog 2 (E. coli) |
| chrX_-_153279697 | 2.35 |
ENST00000444254.1
|
IRAK1
|
interleukin-1 receptor-associated kinase 1 |
| chr4_+_2043777 | 2.34 |
ENST00000409860.1
|
C4orf48
|
chromosome 4 open reading frame 48 |
| chr9_-_140317676 | 2.33 |
ENST00000342129.4
ENST00000340951.4 |
EXD3
|
exonuclease 3'-5' domain containing 3 |
| chr17_-_46671323 | 2.31 |
ENST00000239151.5
|
HOXB5
|
homeobox B5 |
| chr8_+_22446763 | 2.30 |
ENST00000450780.2
ENST00000430850.2 ENST00000447849.1 |
AC037459.4
|
Uncharacterized protein |
| chr2_-_72375167 | 2.30 |
ENST00000001146.2
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1 |
| chr19_+_49496705 | 2.29 |
ENST00000595090.1
|
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr10_+_17272608 | 2.29 |
ENST00000421459.2
|
VIM
|
vimentin |
| chr12_-_53715328 | 2.28 |
ENST00000547757.1
ENST00000394384.3 ENST00000209873.4 |
AAAS
|
achalasia, adrenocortical insufficiency, alacrimia |
| chr7_+_105172612 | 2.28 |
ENST00000493041.1
|
RINT1
|
RAD50 interactor 1 |
| chr2_-_166810261 | 2.28 |
ENST00000243344.7
|
TTC21B
|
tetratricopeptide repeat domain 21B |
| chr9_+_100174344 | 2.28 |
ENST00000422139.2
|
TDRD7
|
tudor domain containing 7 |
| chr4_-_140216948 | 2.27 |
ENST00000265500.4
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr19_-_33793430 | 2.26 |
ENST00000498907.2
|
CEBPA
|
CCAAT/enhancer binding protein (C/EBP), alpha |
| chr9_-_131418944 | 2.26 |
ENST00000419989.1
ENST00000451652.1 ENST00000372715.2 |
WDR34
|
WD repeat domain 34 |
| chr17_-_33446735 | 2.25 |
ENST00000460118.2
ENST00000335858.7 |
RAD51D
|
RAD51 paralog D |
| chr19_-_49339080 | 2.24 |
ENST00000595764.1
|
HSD17B14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr17_-_77813186 | 2.24 |
ENST00000448310.1
ENST00000269397.4 |
CBX4
|
chromobox homolog 4 |
| chr1_-_94374946 | 2.23 |
ENST00000370238.3
|
GCLM
|
glutamate-cysteine ligase, modifier subunit |
| chrX_-_152736013 | 2.20 |
ENST00000330912.2
ENST00000338525.2 ENST00000334497.2 ENST00000370232.1 ENST00000370212.3 ENST00000370211.4 |
TREX2
HAUS7
|
three prime repair exonuclease 2 HAUS augmin-like complex, subunit 7 |
| chr12_-_121410095 | 2.19 |
ENST00000539163.1
|
AC079602.1
|
AC079602.1 |
| chr22_+_23412479 | 2.19 |
ENST00000248996.4
|
GNAZ
|
guanine nucleotide binding protein (G protein), alpha z polypeptide |
| chr17_+_76356516 | 2.19 |
ENST00000592569.1
|
RP11-806H10.4
|
RP11-806H10.4 |
| chr9_+_127539425 | 2.17 |
ENST00000331715.9
|
OLFML2A
|
olfactomedin-like 2A |
| chr7_-_45956856 | 2.16 |
ENST00000428530.1
|
IGFBP3
|
insulin-like growth factor binding protein 3 |
| chr22_+_25003606 | 2.16 |
ENST00000432867.1
|
GGT1
|
gamma-glutamyltransferase 1 |
| chr11_-_44972299 | 2.16 |
ENST00000528473.1
|
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr20_+_37554955 | 2.15 |
ENST00000217429.4
|
FAM83D
|
family with sequence similarity 83, member D |
Network of associatons between targets according to the STRING database.
First level regulatory network of CTCF_CTCFL
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 23.3 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 3.4 | 10.2 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 3.1 | 9.4 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 3.1 | 18.5 | GO:1903412 | response to bile acid(GO:1903412) |
| 3.1 | 9.2 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 1.7 | 30.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 1.3 | 8.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 1.3 | 8.0 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 1.3 | 5.3 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 1.3 | 5.1 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 1.1 | 3.4 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 1.1 | 13.3 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 1.1 | 12.0 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 1.1 | 4.3 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 1.1 | 3.2 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 1.1 | 5.3 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 1.1 | 3.2 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) |
| 1.1 | 1.1 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.9 | 3.6 | GO:0098917 | retrograde trans-synaptic signaling(GO:0098917) |
| 0.9 | 7.3 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.9 | 7.9 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.9 | 4.4 | GO:0090579 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.8 | 3.4 | GO:0035712 | T-helper 2 cell activation(GO:0035712) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.8 | 2.5 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.8 | 2.4 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.8 | 4.7 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.7 | 6.0 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.7 | 3.6 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.7 | 4.4 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.7 | 7.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.7 | 6.4 | GO:0003010 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.7 | 5.0 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.7 | 6.3 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.7 | 2.1 | GO:0034059 | response to anoxia(GO:0034059) positive regulation of penile erection(GO:0060406) |
| 0.7 | 4.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.7 | 2.0 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.6 | 3.9 | GO:0019541 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.6 | 1.9 | GO:0035854 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.6 | 3.9 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.6 | 7.7 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.6 | 2.4 | GO:1904387 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.6 | 2.4 | GO:0042414 | regulation of iron ion transport(GO:0034756) regulation of iron ion transmembrane transport(GO:0034759) epinephrine metabolic process(GO:0042414) |
| 0.6 | 4.2 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.6 | 1.8 | GO:0015709 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.6 | 4.7 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.6 | 1.7 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.6 | 1.7 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.6 | 10.3 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.6 | 2.3 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.6 | 8.9 | GO:0030238 | male sex determination(GO:0030238) |
| 0.6 | 8.9 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.5 | 4.8 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.5 | 17.5 | GO:0036109 | alpha-linolenic acid metabolic process(GO:0036109) linoleic acid metabolic process(GO:0043651) |
| 0.5 | 1.6 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.5 | 1.6 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.5 | 2.1 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.5 | 3.0 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.5 | 3.3 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.5 | 4.2 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.5 | 1.8 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.5 | 2.7 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.5 | 2.3 | GO:1902228 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 0.5 | 3.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.4 | 3.5 | GO:0097461 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.4 | 2.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.4 | 0.4 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.4 | 2.0 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.4 | 2.8 | GO:0046104 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.4 | 1.2 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.4 | 1.5 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.4 | 1.9 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.4 | 4.8 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.4 | 10.3 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.4 | 1.4 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.3 | 4.5 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.3 | 7.6 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.3 | 8.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.3 | 1.7 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.3 | 1.3 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.3 | 4.7 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.3 | 1.7 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.3 | 3.7 | GO:0071316 | cellular response to nicotine(GO:0071316) |
| 0.3 | 1.5 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.3 | 5.4 | GO:0039532 | negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039532) |
| 0.3 | 2.4 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.3 | 1.2 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.3 | 5.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.3 | 1.4 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.3 | 4.5 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.3 | 0.9 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.3 | 1.1 | GO:0072302 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.3 | 2.3 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.3 | 1.1 | GO:0018283 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.3 | 0.8 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.3 | 1.6 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.3 | 5.7 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.3 | 6.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.3 | 0.5 | GO:0045720 | negative regulation of integrin biosynthetic process(GO:0045720) |
| 0.3 | 1.3 | GO:1901536 | negative regulation of DNA demethylation(GO:1901536) |
| 0.3 | 1.8 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.3 | 1.8 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.3 | 5.5 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.2 | 11.4 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.2 | 2.9 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.2 | 9.9 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.2 | 1.7 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.2 | 3.8 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.2 | 3.3 | GO:0032308 | regulation of prostaglandin secretion(GO:0032306) positive regulation of prostaglandin secretion(GO:0032308) |
| 0.2 | 1.9 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.2 | 1.6 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.2 | 3.0 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.2 | 0.7 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.2 | 0.9 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 0.2 | 6.3 | GO:0035791 | platelet-derived growth factor receptor-beta signaling pathway(GO:0035791) |
| 0.2 | 0.4 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.2 | 1.0 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.2 | 1.2 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) L-kynurenine metabolic process(GO:0097052) |
| 0.2 | 20.4 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.2 | 1.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.2 | 3.1 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.2 | 1.0 | GO:0006490 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.2 | 2.3 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.2 | 5.4 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.2 | 1.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 1.8 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.2 | 1.2 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.2 | 0.2 | GO:0046440 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.2 | 0.7 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.2 | 1.2 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.2 | 9.3 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.2 | 3.7 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.2 | 0.8 | GO:0009149 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) |
| 0.2 | 0.8 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.2 | 1.9 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.2 | 3.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 0.6 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.2 | 0.5 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.2 | 0.9 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.2 | 0.6 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.2 | 3.6 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.1 | 1.5 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.3 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.1 | 0.6 | GO:0070845 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.1 | 0.7 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 0.7 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.1 | 0.7 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.1 | 0.7 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.1 | 4.3 | GO:0060065 | uterus development(GO:0060065) |
| 0.1 | 4.7 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.1 | 2.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 4.6 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.1 | 0.3 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.1 | 0.7 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 | 0.4 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 11.1 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.1 | 0.9 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 1.3 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.1 | 0.3 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.1 | 0.9 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.1 | 2.4 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 1.9 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.1 | 1.5 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 1.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.1 | 3.7 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 4.9 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.1 | 1.6 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 2.4 | GO:0051775 | response to redox state(GO:0051775) |
| 0.1 | 0.4 | GO:0034130 | toll-like receptor 1 signaling pathway(GO:0034130) |
| 0.1 | 0.4 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.1 | 2.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.7 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 1.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.6 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.1 | 0.5 | GO:0015942 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) formate metabolic process(GO:0015942) |
| 0.1 | 0.8 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.1 | 2.6 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.1 | 1.0 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 0.3 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.1 | 0.7 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.1 | 1.2 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.1 | 2.5 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 4.3 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.1 | 1.6 | GO:1903351 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.1 | 0.9 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.5 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 0.1 | 1.0 | GO:0050808 | synapse organization(GO:0050808) |
| 0.1 | 3.1 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.1 | 0.7 | GO:0035087 | siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.1 | 1.7 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.4 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.1 | 1.6 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 0.6 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 5.6 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.1 | 0.3 | GO:1900276 | negative regulation of phospholipase C activity(GO:1900275) regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.1 | 4.8 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.1 | 1.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 1.6 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.1 | 2.3 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.6 | GO:0060283 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.4 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 0.3 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.1 | 0.4 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 1.5 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.1 | 1.5 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.1 | 0.8 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 0.2 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.1 | 0.4 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.1 | 1.7 | GO:0021756 | striatum development(GO:0021756) |
| 0.1 | 5.1 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.1 | 1.1 | GO:1902074 | response to salt(GO:1902074) |
| 0.1 | 3.5 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.1 | 0.9 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.3 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.1 | 0.8 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 1.0 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.1 | 2.9 | GO:0035825 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.1 | 0.3 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.2 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.1 | 0.3 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 1.0 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.8 | GO:0018202 | peptidyl-histidine modification(GO:0018202) |
| 0.1 | 0.6 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 4.0 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.1 | 1.6 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.1 | 1.3 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.1 | 0.4 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.8 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 5.9 | GO:0051439 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) |
| 0.1 | 5.9 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.1 | 0.9 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.1 | 1.5 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.1 | 1.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 2.1 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.3 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.8 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.1 | 3.3 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.1 | 0.8 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 0.2 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.1 | 1.0 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 2.0 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 1.1 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 2.2 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 1.5 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 10.7 | GO:0051384 | response to glucocorticoid(GO:0051384) |
| 0.1 | 0.2 | GO:1900248 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.1 | 1.8 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 1.9 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 0.4 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.1 | 3.2 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.1 | 1.1 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 0.4 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 1.0 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 4.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 2.5 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 0.4 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.1 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.0 | 2.5 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.2 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.5 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.0 | 0.7 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.4 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 4.5 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.2 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.5 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.3 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 1.5 | GO:0038202 | TORC1 signaling(GO:0038202) |
| 0.0 | 0.1 | GO:0042713 | sperm ejaculation(GO:0042713) |
| 0.0 | 0.8 | GO:0009125 | nucleoside monophosphate catabolic process(GO:0009125) |
| 0.0 | 2.3 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 3.9 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 6.1 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 1.6 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.5 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.0 | 4.7 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.6 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.4 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.7 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 1.2 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.8 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 | 1.1 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.7 | GO:0014898 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.9 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 0.2 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.1 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 1.4 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.2 | GO:0006273 | lagging strand elongation(GO:0006273) |
| 0.0 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.4 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.2 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.0 | 0.7 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.9 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.5 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.9 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 1.4 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 0.4 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 2.0 | GO:0040014 | regulation of multicellular organism growth(GO:0040014) |
| 0.0 | 0.8 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 4.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.6 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 2.5 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 0.8 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.1 | GO:1903027 | regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.8 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 2.7 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 2.1 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 4.7 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.0 | 0.3 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 4.1 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.3 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.5 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 1.9 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.1 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.0 | 1.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.4 | GO:0046602 | mitotic centrosome separation(GO:0007100) regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 2.9 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.0 | 0.1 | GO:0006424 | glutamyl-tRNA aminoacylation(GO:0006424) |
| 0.0 | 0.6 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.6 | GO:0009988 | cell-cell recognition(GO:0009988) |
| 0.0 | 0.7 | GO:0006536 | glutamate metabolic process(GO:0006536) |
| 0.0 | 0.5 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 1.0 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.7 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.7 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.4 | GO:0050687 | negative regulation of defense response to virus(GO:0050687) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.4 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 1.4 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.2 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.8 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.1 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.8 | GO:0070303 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 2.4 | GO:0009566 | fertilization(GO:0009566) |
| 0.0 | 0.1 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.0 | 0.4 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.5 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 1.7 | GO:0050709 | negative regulation of protein secretion(GO:0050709) |
| 0.0 | 0.5 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.7 | GO:0001947 | heart looping(GO:0001947) |
| 0.0 | 0.1 | GO:0055074 | calcium ion homeostasis(GO:0055074) |
| 0.0 | 0.3 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.1 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.2 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.9 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 1.0 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.0 | 1.2 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 1.2 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.0 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.4 | GO:0035272 | exocrine system development(GO:0035272) |
| 0.0 | 0.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.1 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 0.3 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.3 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 0.3 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.6 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 15.8 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 2.1 | 23.0 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 1.2 | 5.0 | GO:0000811 | GINS complex(GO:0000811) |
| 1.2 | 12.0 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 1.2 | 7.2 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 1.0 | 10.5 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.9 | 4.3 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.8 | 2.4 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.7 | 5.9 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.7 | 8.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.6 | 8.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.6 | 2.4 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.6 | 4.8 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.6 | 2.4 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.6 | 1.7 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.6 | 2.9 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.6 | 4.5 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.6 | 5.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.5 | 11.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.5 | 5.6 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.5 | 3.0 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.5 | 3.7 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.5 | 3.6 | GO:0043196 | varicosity(GO:0043196) |
| 0.4 | 1.3 | GO:0035101 | FACT complex(GO:0035101) |
| 0.4 | 3.0 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.4 | 4.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.4 | 3.9 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.4 | 5.9 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.4 | 2.3 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.4 | 4.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.4 | 1.5 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.4 | 1.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.4 | 1.1 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.4 | 1.1 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.3 | 2.3 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.3 | 21.1 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.3 | 0.8 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.3 | 6.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.3 | 4.7 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.3 | 4.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 5.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 3.6 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.2 | 4.7 | GO:0032059 | bleb(GO:0032059) |
| 0.2 | 7.9 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.2 | 9.3 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.2 | 1.0 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.2 | 0.8 | GO:0031523 | Myb complex(GO:0031523) |
| 0.2 | 4.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.2 | 4.4 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.2 | 1.4 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.2 | 7.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.2 | 0.9 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.2 | 2.7 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.2 | 1.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.2 | 2.3 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.2 | 1.2 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.2 | 6.7 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.2 | 3.7 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.2 | 1.7 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.2 | 1.7 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.2 | 2.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 1.2 | GO:0097361 | CIA complex(GO:0097361) |
| 0.2 | 1.5 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.7 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.1 | 1.0 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 1.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 1.2 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 5.0 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.4 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
| 0.1 | 2.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 1.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.6 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 3.3 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 0.9 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 3.6 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 1.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 0.5 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.1 | 3.3 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 0.7 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 1.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 1.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.5 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 4.7 | GO:0000791 | euchromatin(GO:0000791) |
| 0.1 | 11.0 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 0.6 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.4 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.1 | 9.8 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 17.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 0.8 | GO:0001939 | female pronucleus(GO:0001939) male pronucleus(GO:0001940) |
| 0.1 | 0.2 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.1 | 0.4 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.3 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.1 | 5.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.8 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 5.4 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 0.4 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 10.1 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.1 | 2.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 4.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 3.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 7.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 1.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 5.0 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 1.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 1.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 1.0 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 1.3 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 1.5 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 0.9 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.9 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 5.5 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.6 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.4 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 1.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 2.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.1 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.0 | 0.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 1.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.9 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 3.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 1.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 56.8 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 10.4 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.3 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.0 | 0.8 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 2.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 2.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 2.0 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.7 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 10.5 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 1.8 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 4.7 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.8 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.2 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.7 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 0.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.6 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 10.4 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.4 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 3.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.2 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 2.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.3 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 4.9 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 1.7 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.3 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 3.5 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 0.1 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.7 | GO:0030496 | midbody(GO:0030496) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.4 | GO:0045485 | omega-6 fatty acid desaturase activity(GO:0045485) |
| 3.4 | 10.2 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 2.4 | 12.0 | GO:0003947 | (N-acetylneuraminyl)-galactosylglucosylceramide N-acetylgalactosaminyltransferase activity(GO:0003947) |
| 2.1 | 6.3 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 1.8 | 5.4 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 1.8 | 7.0 | GO:0005019 | platelet-derived growth factor beta-receptor activity(GO:0005019) |
| 1.6 | 4.8 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 1.5 | 18.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 1.5 | 8.8 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 1.4 | 4.2 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 1.3 | 3.8 | GO:0004513 | neolactotetraosylceramide alpha-2,3-sialyltransferase activity(GO:0004513) lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 1.2 | 4.9 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 1.1 | 11.2 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 1.1 | 3.2 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 1.0 | 3.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 1.0 | 7.7 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 1.0 | 2.9 | GO:0048257 | 3'-flap endonuclease activity(GO:0048257) |
| 0.9 | 9.7 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.9 | 6.0 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.8 | 7.2 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.8 | 4.7 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.7 | 2.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.7 | 4.1 | GO:0016213 | linoleoyl-CoA desaturase activity(GO:0016213) |
| 0.7 | 2.0 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.6 | 5.8 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.6 | 2.4 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.6 | 2.4 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) copper-dependent protein binding(GO:0032767) |
| 0.6 | 3.0 | GO:0098626 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.6 | 2.4 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.6 | 1.8 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.6 | 10.3 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.6 | 1.1 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.6 | 5.0 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.5 | 4.4 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.5 | 2.1 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.5 | 28.8 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.5 | 2.0 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.5 | 3.5 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.5 | 2.4 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 0.5 | 3.9 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.5 | 2.8 | GO:0004797 | thymidine kinase activity(GO:0004797) |
| 0.5 | 6.4 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.5 | 4.5 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.4 | 6.6 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.4 | 8.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.4 | 1.2 | GO:0047536 | 2-aminoadipate transaminase activity(GO:0047536) |
| 0.4 | 3.7 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.4 | 4.0 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.4 | 1.2 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.4 | 4.5 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.4 | 1.5 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.4 | 12.0 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.4 | 1.1 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.3 | 1.0 | GO:0052830 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.3 | 7.8 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.3 | 4.7 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.3 | 2.3 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.3 | 5.6 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.3 | 13.6 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.3 | 2.2 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.3 | 5.0 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.3 | 0.9 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.3 | 3.0 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.3 | 2.3 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.3 | 5.0 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.3 | 3.3 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.3 | 1.1 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.3 | 9.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.3 | 9.9 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.3 | 2.9 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.3 | 3.0 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 1.5 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.2 | 0.9 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 0.9 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.2 | 7.2 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.2 | 1.1 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.2 | 0.9 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.2 | 2.7 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.2 | 6.5 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.2 | 0.7 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.2 | 3.1 | GO:0089720 | caspase binding(GO:0089720) |
| 0.2 | 0.9 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.2 | 0.9 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitrite reductase activity(GO:0098809) |
| 0.2 | 0.6 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.2 | 1.7 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.2 | 1.3 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.2 | 1.2 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.2 | 1.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.2 | 9.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 3.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 11.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 11.1 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.2 | 3.8 | GO:0008061 | chitin binding(GO:0008061) |
| 0.2 | 5.7 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.2 | 1.5 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.2 | 1.7 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.2 | 7.7 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.2 | 4.5 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.2 | 1.2 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.2 | 5.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 2.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.2 | 2.5 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.2 | 0.7 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.2 | 2.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.2 | 1.8 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.2 | 0.8 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.2 | 2.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.2 | 0.5 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.2 | 1.5 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 2.5 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.1 | 0.4 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 1.7 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 1.3 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 0.9 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.1 | 4.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 1.7 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 1.2 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.1 | 1.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.4 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.1 | 1.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 2.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 8.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 0.8 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.1 | 2.0 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 1.6 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.9 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.4 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 0.9 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.5 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 1.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.5 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.1 | 14.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 2.0 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.8 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.1 | 0.9 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.7 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.1 | 1.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 4.7 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 0.4 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) |
| 0.1 | 3.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 3.2 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 1.0 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.1 | 2.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 2.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.7 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.4 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 1.2 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.1 | 1.9 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.1 | 2.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 2.2 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 0.6 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.1 | 1.0 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 1.4 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 12.2 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.1 | 4.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.2 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 1.9 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 0.2 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.1 | 2.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.3 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.1 | 4.3 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 0.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 12.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 1.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 25.5 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 0.7 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 5.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.8 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 0.2 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.1 | 1.0 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 3.7 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.1 | 1.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 1.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.9 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 2.4 | GO:0044390 | ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.6 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.2 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 1.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.3 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 1.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.2 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 1.1 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.9 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 1.2 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 5.3 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.9 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 1.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.7 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.3 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.2 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 3.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.3 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0030284 | mineralocorticoid receptor activity(GO:0017082) estrogen receptor activity(GO:0030284) |
| 0.0 | 1.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.0 | 0.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 3.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.2 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 1.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 1.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 3.2 | GO:0005230 | extracellular ligand-gated ion channel activity(GO:0005230) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.5 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0004818 | glutamate-tRNA ligase activity(GO:0004818) |
| 0.0 | 0.2 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.8 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.4 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 1.0 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.8 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.2 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.8 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.3 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.2 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 13.8 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.0 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 0.1 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.8 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.1 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.0 | 0.7 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 2.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.4 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 3.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 2.5 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.7 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.8 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.6 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.9 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.2 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.5 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 2.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 21.9 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.4 | 9.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.3 | 4.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.3 | 9.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.3 | 22.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 17.1 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.2 | 9.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 11.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 16.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.2 | 18.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 9.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 2.9 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 4.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 5.7 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.1 | 8.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 7.0 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 3.1 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.1 | 2.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 0.7 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.1 | 3.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 2.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 5.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 0.9 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 8.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 5.2 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 2.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 2.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.8 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 2.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 3.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.5 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 3.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.0 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.8 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.8 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 2.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.5 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 1.4 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 2.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.9 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.0 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.8 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.3 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 1.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.8 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.6 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 4.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 4.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 18.5 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 1.0 | 24.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.7 | 19.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.6 | 17.5 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.5 | 7.7 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.3 | 15.3 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.3 | 8.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.3 | 4.3 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.3 | 4.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.3 | 9.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.3 | 12.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 5.0 | REACTOME DNA STRAND ELONGATION | Genes involved in DNA strand elongation |
| 0.2 | 4.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 8.4 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.2 | 3.9 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 12.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.2 | 6.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 2.0 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.2 | 3.9 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 3.5 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.2 | 14.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 0.9 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.2 | 4.7 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 1.7 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 9.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 2.8 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 3.4 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 8.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 2.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 2.0 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 2.0 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 5.2 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.1 | 1.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 3.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 4.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 2.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 2.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 2.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 8.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 1.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 4.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 5.5 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 1.9 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 2.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.2 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.0 | 1.7 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 1.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.5 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 4.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.5 | REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | Genes involved in Cleavage of Growing Transcript in the Termination Region |
| 0.0 | 6.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 3.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.9 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 1.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 1.2 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 1.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 3.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.6 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 1.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 1.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 2.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 5.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 2.2 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.8 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.5 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.0 | 1.0 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.2 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.7 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 1.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 0.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 1.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.5 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 MUTANTS | Genes involved in Signaling by FGFR1 mutants |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |