Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
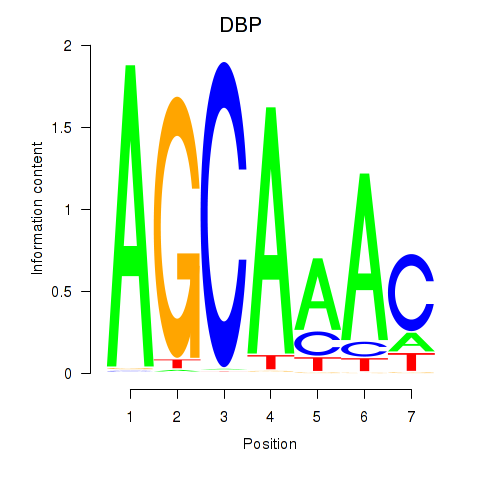
Results for DBP
Z-value: 0.63
Transcription factors associated with DBP
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DBP
|
ENSG00000105516.6 | D-box binding PAR bZIP transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DBP | hg19_v2_chr19_-_49140692_49140709 | 0.78 | 4.7e-05 | Click! |
Activity profile of DBP motif
Sorted Z-values of DBP motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_56492989 | 0.60 |
ENST00000583753.1
|
RNF43
|
ring finger protein 43 |
| chr2_-_216257849 | 0.57 |
ENST00000456923.1
|
FN1
|
fibronectin 1 |
| chr21_+_17791648 | 0.55 |
ENST00000602892.1
ENST00000418813.2 ENST00000435697.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chrX_+_23801280 | 0.55 |
ENST00000379251.3
ENST00000379253.3 ENST00000379254.1 ENST00000379270.4 |
SAT1
|
spermidine/spermine N1-acetyltransferase 1 |
| chr12_-_89746173 | 0.52 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr4_-_176733897 | 0.52 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A |
| chr17_+_42429493 | 0.51 |
ENST00000586242.1
|
GRN
|
granulin |
| chr4_-_23891693 | 0.50 |
ENST00000264867.2
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr21_-_28215332 | 0.47 |
ENST00000517777.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr10_-_64576105 | 0.47 |
ENST00000242480.3
ENST00000411732.1 |
EGR2
|
early growth response 2 |
| chr7_-_150946015 | 0.45 |
ENST00000262188.8
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr2_+_210444748 | 0.45 |
ENST00000392194.1
|
MAP2
|
microtubule-associated protein 2 |
| chr2_+_210444142 | 0.45 |
ENST00000360351.4
ENST00000361559.4 |
MAP2
|
microtubule-associated protein 2 |
| chr19_+_11455900 | 0.44 |
ENST00000588790.1
|
CCDC159
|
coiled-coil domain containing 159 |
| chr7_-_41740181 | 0.43 |
ENST00000442711.1
|
INHBA
|
inhibin, beta A |
| chr2_+_182850743 | 0.42 |
ENST00000409702.1
|
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr3_-_185538849 | 0.41 |
ENST00000421047.2
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr5_-_95550754 | 0.40 |
ENST00000502437.1
|
RP11-254I22.3
|
RP11-254I22.3 |
| chr16_-_79634595 | 0.40 |
ENST00000326043.4
ENST00000393350.1 |
MAF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr14_-_23652849 | 0.40 |
ENST00000316902.7
ENST00000469263.1 ENST00000525062.1 ENST00000524758.1 |
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr1_+_114522049 | 0.39 |
ENST00000369551.1
ENST00000320334.4 |
OLFML3
|
olfactomedin-like 3 |
| chr7_-_121784285 | 0.39 |
ENST00000417368.2
|
AASS
|
aminoadipate-semialdehyde synthase |
| chr12_-_6483969 | 0.39 |
ENST00000396966.2
|
SCNN1A
|
sodium channel, non-voltage-gated 1 alpha subunit |
| chr16_+_30675654 | 0.38 |
ENST00000287468.5
ENST00000395073.2 |
FBRS
|
fibrosin |
| chr3_-_48471454 | 0.38 |
ENST00000296440.6
ENST00000448774.2 |
PLXNB1
|
plexin B1 |
| chr1_+_104104379 | 0.37 |
ENST00000435302.1
|
AMY2B
|
amylase, alpha 2B (pancreatic) |
| chr1_+_82266053 | 0.36 |
ENST00000370715.1
ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2
|
latrophilin 2 |
| chr8_-_101718991 | 0.36 |
ENST00000517990.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr1_+_155051305 | 0.35 |
ENST00000368408.3
|
EFNA3
|
ephrin-A3 |
| chr1_+_61547894 | 0.34 |
ENST00000403491.3
|
NFIA
|
nuclear factor I/A |
| chr5_-_27038683 | 0.34 |
ENST00000511822.1
ENST00000231021.4 |
CDH9
|
cadherin 9, type 2 (T1-cadherin) |
| chrX_+_134654540 | 0.34 |
ENST00000370752.4
|
DDX26B
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chr3_-_47023455 | 0.34 |
ENST00000446836.1
ENST00000425441.1 |
CCDC12
|
coiled-coil domain containing 12 |
| chr8_-_22550815 | 0.33 |
ENST00000317216.2
|
EGR3
|
early growth response 3 |
| chr6_+_33168597 | 0.32 |
ENST00000374675.3
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr14_-_37051798 | 0.32 |
ENST00000258829.5
|
NKX2-8
|
NK2 homeobox 8 |
| chr2_+_210517895 | 0.32 |
ENST00000447185.1
|
MAP2
|
microtubule-associated protein 2 |
| chr11_+_64009072 | 0.32 |
ENST00000535135.1
ENST00000394540.3 |
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr6_-_33168391 | 0.31 |
ENST00000374685.4
ENST00000413614.2 ENST00000374680.3 |
RXRB
|
retinoid X receptor, beta |
| chr16_+_30960375 | 0.31 |
ENST00000318663.4
ENST00000566237.1 ENST00000562699.1 |
ORAI3
|
ORAI calcium release-activated calcium modulator 3 |
| chr2_+_210518057 | 0.30 |
ENST00000452717.1
|
MAP2
|
microtubule-associated protein 2 |
| chr1_+_12538594 | 0.29 |
ENST00000543710.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr2_+_171571827 | 0.28 |
ENST00000375281.3
|
SP5
|
Sp5 transcription factor |
| chr10_+_24755416 | 0.28 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr12_-_92536433 | 0.28 |
ENST00000551563.2
ENST00000546975.1 ENST00000549802.1 |
C12orf79
|
chromosome 12 open reading frame 79 |
| chr6_+_26251835 | 0.28 |
ENST00000356350.2
|
HIST1H2BH
|
histone cluster 1, H2bh |
| chr14_-_73360796 | 0.28 |
ENST00000556509.1
ENST00000541685.1 ENST00000546183.1 |
DPF3
|
D4, zinc and double PHD fingers, family 3 |
| chr12_-_99548524 | 0.28 |
ENST00000549558.2
ENST00000550693.2 ENST00000549493.2 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr19_+_41856816 | 0.28 |
ENST00000539627.1
|
TMEM91
|
transmembrane protein 91 |
| chr14_+_29236269 | 0.27 |
ENST00000313071.4
|
FOXG1
|
forkhead box G1 |
| chr16_+_2059872 | 0.27 |
ENST00000567649.1
|
NPW
|
neuropeptide W |
| chr1_-_40367668 | 0.27 |
ENST00000397332.2
ENST00000429311.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr11_-_46142948 | 0.27 |
ENST00000257821.4
|
PHF21A
|
PHD finger protein 21A |
| chr7_+_114055052 | 0.27 |
ENST00000462331.1
ENST00000408937.3 ENST00000403559.4 ENST00000350908.4 ENST00000393498.2 ENST00000393495.3 ENST00000378237.3 ENST00000393489.3 |
FOXP2
|
forkhead box P2 |
| chr8_+_42195972 | 0.26 |
ENST00000532157.1
ENST00000265421.4 ENST00000520008.1 |
POLB
|
polymerase (DNA directed), beta |
| chr6_-_94129244 | 0.26 |
ENST00000369303.4
ENST00000369297.1 |
EPHA7
|
EPH receptor A7 |
| chr6_+_33168637 | 0.26 |
ENST00000374677.3
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr6_+_33168189 | 0.26 |
ENST00000444757.1
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7 |
| chrX_-_151619746 | 0.26 |
ENST00000370314.4
|
GABRA3
|
gamma-aminobutyric acid (GABA) A receptor, alpha 3 |
| chr17_+_2240775 | 0.26 |
ENST00000268989.3
ENST00000426855.2 |
SGSM2
|
small G protein signaling modulator 2 |
| chr2_-_118943930 | 0.25 |
ENST00000449075.1
ENST00000414886.1 ENST00000449819.1 |
AC093901.1
|
AC093901.1 |
| chr16_-_18468926 | 0.25 |
ENST00000545114.1
|
RP11-1212A22.4
|
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr2_+_210444298 | 0.25 |
ENST00000445941.1
|
MAP2
|
microtubule-associated protein 2 |
| chr12_-_89746264 | 0.24 |
ENST00000548755.1
|
DUSP6
|
dual specificity phosphatase 6 |
| chr16_+_23194033 | 0.24 |
ENST00000300061.2
|
SCNN1G
|
sodium channel, non-voltage-gated 1, gamma subunit |
| chr9_-_89562104 | 0.24 |
ENST00000298743.7
|
GAS1
|
growth arrest-specific 1 |
| chr2_+_105050794 | 0.23 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr6_+_88054530 | 0.22 |
ENST00000388923.4
|
C6orf163
|
chromosome 6 open reading frame 163 |
| chr1_+_24645807 | 0.22 |
ENST00000361548.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr19_-_48894104 | 0.22 |
ENST00000597017.1
|
KDELR1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr19_+_13135386 | 0.22 |
ENST00000360105.4
ENST00000588228.1 ENST00000591028.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr14_+_23654525 | 0.22 |
ENST00000399910.1
ENST00000492621.1 |
C14orf164
|
chromosome 14 open reading frame 164 |
| chr5_+_179921430 | 0.22 |
ENST00000393356.1
|
CNOT6
|
CCR4-NOT transcription complex, subunit 6 |
| chr1_+_24646002 | 0.21 |
ENST00000356046.2
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr21_+_17791838 | 0.21 |
ENST00000453910.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr1_+_24645865 | 0.21 |
ENST00000342072.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr19_+_36208877 | 0.21 |
ENST00000420124.1
ENST00000222270.7 ENST00000341701.1 |
KMT2B
|
Histone-lysine N-methyltransferase 2B |
| chr6_+_45296048 | 0.21 |
ENST00000465038.2
ENST00000352853.5 ENST00000541979.1 ENST00000371438.1 |
RUNX2
|
runt-related transcription factor 2 |
| chr3_+_184098065 | 0.21 |
ENST00000348986.3
|
CHRD
|
chordin |
| chr3_+_184097836 | 0.21 |
ENST00000204604.1
ENST00000310236.3 |
CHRD
|
chordin |
| chr1_+_223101757 | 0.20 |
ENST00000284476.6
|
DISP1
|
dispatched homolog 1 (Drosophila) |
| chr1_+_82165350 | 0.20 |
ENST00000359929.3
|
LPHN2
|
latrophilin 2 |
| chr3_+_184097905 | 0.20 |
ENST00000450923.1
|
CHRD
|
chordin |
| chr6_+_31514622 | 0.19 |
ENST00000376146.4
|
NFKBIL1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
| chr3_+_72201910 | 0.19 |
ENST00000469178.1
ENST00000485404.1 |
LINC00870
|
long intergenic non-protein coding RNA 870 |
| chr9_+_35829208 | 0.19 |
ENST00000439587.2
ENST00000377991.4 |
TMEM8B
|
transmembrane protein 8B |
| chr1_+_211433275 | 0.19 |
ENST00000367005.4
|
RCOR3
|
REST corepressor 3 |
| chr14_-_36988882 | 0.18 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr4_+_71587669 | 0.18 |
ENST00000381006.3
ENST00000226328.4 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr4_-_186732048 | 0.18 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr13_+_44947941 | 0.18 |
ENST00000379179.3
|
SERP2
|
stress-associated endoplasmic reticulum protein family member 2 |
| chr1_-_92952433 | 0.18 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr7_+_145813453 | 0.18 |
ENST00000361727.3
|
CNTNAP2
|
contactin associated protein-like 2 |
| chr2_-_60780607 | 0.18 |
ENST00000537768.1
ENST00000335712.6 ENST00000356842.4 |
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chr5_+_140588269 | 0.18 |
ENST00000541609.1
ENST00000239450.2 |
PCDHB12
|
protocadherin beta 12 |
| chr1_+_104293028 | 0.18 |
ENST00000370079.3
|
AMY1C
|
amylase, alpha 1C (salivary) |
| chr3_-_114343768 | 0.17 |
ENST00000393785.2
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr2_+_234602305 | 0.17 |
ENST00000406651.1
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr14_+_76452090 | 0.17 |
ENST00000314067.6
ENST00000238628.6 ENST00000556742.1 |
IFT43
|
intraflagellar transport 43 homolog (Chlamydomonas) |
| chr8_+_85095769 | 0.17 |
ENST00000518566.1
|
RALYL
|
RALY RNA binding protein-like |
| chr14_-_23479331 | 0.17 |
ENST00000397377.1
ENST00000397379.3 ENST00000406429.2 ENST00000341470.4 ENST00000555998.1 ENST00000397376.2 ENST00000553675.1 ENST00000553931.1 ENST00000555575.1 ENST00000553958.1 ENST00000555098.1 ENST00000556419.1 ENST00000553606.1 ENST00000299088.6 ENST00000554179.1 ENST00000397382.4 |
C14orf93
|
chromosome 14 open reading frame 93 |
| chr7_+_139529040 | 0.17 |
ENST00000455353.1
ENST00000458722.1 ENST00000411653.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr1_+_104159999 | 0.17 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr1_+_24645915 | 0.17 |
ENST00000350501.5
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr4_-_87281224 | 0.17 |
ENST00000395169.3
ENST00000395161.2 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr7_+_103969104 | 0.17 |
ENST00000424859.1
ENST00000535008.1 ENST00000401970.2 ENST00000543266.1 |
LHFPL3
|
lipoma HMGIC fusion partner-like 3 |
| chr18_-_51751132 | 0.17 |
ENST00000256429.3
|
MBD2
|
methyl-CpG binding domain protein 2 |
| chr5_+_179921344 | 0.17 |
ENST00000261951.4
|
CNOT6
|
CCR4-NOT transcription complex, subunit 6 |
| chr5_-_45696253 | 0.16 |
ENST00000303230.4
|
HCN1
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 1 |
| chr2_+_169659121 | 0.16 |
ENST00000397206.2
ENST00000397209.2 ENST00000421711.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr5_+_129083772 | 0.16 |
ENST00000564719.1
|
KIAA1024L
|
KIAA1024-like |
| chr8_+_85095553 | 0.16 |
ENST00000521268.1
|
RALYL
|
RALY RNA binding protein-like |
| chr7_-_37026108 | 0.16 |
ENST00000396045.3
|
ELMO1
|
engulfment and cell motility 1 |
| chr8_-_22549856 | 0.16 |
ENST00000522910.1
|
EGR3
|
early growth response 3 |
| chr5_+_50679506 | 0.15 |
ENST00000511384.1
|
ISL1
|
ISL LIM homeobox 1 |
| chr17_-_8263538 | 0.15 |
ENST00000535173.1
|
AC135178.1
|
HCG1985372; Uncharacterized protein; cDNA FLJ37541 fis, clone BRCAN2026340 |
| chr9_+_84304628 | 0.15 |
ENST00000437181.1
|
RP11-154D17.1
|
RP11-154D17.1 |
| chrX_+_107288280 | 0.15 |
ENST00000458383.1
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr4_-_100242549 | 0.15 |
ENST00000305046.8
ENST00000394887.3 |
ADH1B
|
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr7_+_142374104 | 0.15 |
ENST00000604952.1
|
MTRNR2L6
|
MT-RNR2-like 6 |
| chr12_-_58212487 | 0.15 |
ENST00000549994.1
|
AVIL
|
advillin |
| chr2_-_60780702 | 0.14 |
ENST00000359629.5
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chr3_+_155838337 | 0.14 |
ENST00000490337.1
ENST00000389636.5 |
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr8_-_101719159 | 0.14 |
ENST00000520868.1
ENST00000522658.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr7_+_13141097 | 0.14 |
ENST00000411542.1
|
AC011288.2
|
AC011288.2 |
| chrX_+_49028265 | 0.14 |
ENST00000376322.3
ENST00000376327.5 |
PLP2
|
proteolipid protein 2 (colonic epithelium-enriched) |
| chr10_-_69991865 | 0.14 |
ENST00000373673.3
|
ATOH7
|
atonal homolog 7 (Drosophila) |
| chr1_+_155051379 | 0.14 |
ENST00000418360.2
|
EFNA3
|
ephrin-A3 |
| chr4_-_184243561 | 0.14 |
ENST00000514470.1
ENST00000541814.1 |
CLDN24
|
claudin 24 |
| chr11_-_94706705 | 0.14 |
ENST00000279839.6
|
CWC15
|
CWC15 spliceosome-associated protein homolog (S. cerevisiae) |
| chr17_+_2240916 | 0.14 |
ENST00000574563.1
|
SGSM2
|
small G protein signaling modulator 2 |
| chr2_-_60780546 | 0.14 |
ENST00000358510.4
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chr10_+_60936921 | 0.14 |
ENST00000373878.3
|
PHYHIPL
|
phytanoyl-CoA 2-hydroxylase interacting protein-like |
| chr4_-_87281196 | 0.14 |
ENST00000359221.3
|
MAPK10
|
mitogen-activated protein kinase 10 |
| chr6_-_75953484 | 0.13 |
ENST00000472311.2
ENST00000460985.1 ENST00000377978.3 ENST00000509698.1 ENST00000230459.4 ENST00000370089.2 |
COX7A2
|
cytochrome c oxidase subunit VIIa polypeptide 2 (liver) |
| chr8_-_22550691 | 0.13 |
ENST00000519492.1
|
EGR3
|
early growth response 3 |
| chr17_+_38498594 | 0.13 |
ENST00000394081.3
|
RARA
|
retinoic acid receptor, alpha |
| chr7_+_139528952 | 0.13 |
ENST00000416849.2
ENST00000436047.2 ENST00000414508.2 ENST00000448866.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr1_+_24646263 | 0.13 |
ENST00000524724.1
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr10_-_50603497 | 0.13 |
ENST00000374139.2
|
DRGX
|
dorsal root ganglia homeobox |
| chr4_+_71588372 | 0.13 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr4_+_117220016 | 0.13 |
ENST00000604093.1
|
MTRNR2L13
|
MT-RNR2-like 13 (pseudogene) |
| chr3_-_50541028 | 0.13 |
ENST00000266039.3
ENST00000435965.1 ENST00000395083.1 |
CACNA2D2
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2 |
| chr8_+_42196000 | 0.13 |
ENST00000518925.1
ENST00000538005.1 |
POLB
|
polymerase (DNA directed), beta |
| chr12_-_99548645 | 0.13 |
ENST00000549025.2
|
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr2_+_168725458 | 0.13 |
ENST00000392690.3
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr1_+_154229547 | 0.13 |
ENST00000428595.1
|
UBAP2L
|
ubiquitin associated protein 2-like |
| chr17_-_27332931 | 0.13 |
ENST00000442608.3
ENST00000335960.6 |
SEZ6
|
seizure related 6 homolog (mouse) |
| chr8_-_72268721 | 0.13 |
ENST00000419131.1
ENST00000388743.2 |
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chrX_-_10851762 | 0.13 |
ENST00000380785.1
ENST00000380787.1 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chrY_+_4868267 | 0.13 |
ENST00000333703.4
|
PCDH11Y
|
protocadherin 11 Y-linked |
| chr8_+_99076750 | 0.12 |
ENST00000545282.1
|
C8orf47
|
chromosome 8 open reading frame 47 |
| chr5_-_892880 | 0.12 |
ENST00000467963.1
|
BRD9
|
bromodomain containing 9 |
| chr2_-_60780536 | 0.12 |
ENST00000538214.1
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chrX_+_107288239 | 0.12 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr18_-_35145981 | 0.12 |
ENST00000420428.2
ENST00000412753.1 |
CELF4
|
CUGBP, Elav-like family member 4 |
| chr20_+_44650348 | 0.12 |
ENST00000454036.2
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5 |
| chr6_-_31514516 | 0.12 |
ENST00000303892.5
ENST00000483251.1 |
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr8_-_6420759 | 0.12 |
ENST00000523120.1
|
ANGPT2
|
angiopoietin 2 |
| chr4_+_185395947 | 0.12 |
ENST00000605834.1
|
RP11-326I11.3
|
RP11-326I11.3 |
| chr16_+_67022582 | 0.12 |
ENST00000541479.1
ENST00000338718.4 |
CES4A
|
carboxylesterase 4A |
| chr16_+_67022633 | 0.12 |
ENST00000398354.1
ENST00000326686.5 |
CES4A
|
carboxylesterase 4A |
| chr6_-_117150198 | 0.12 |
ENST00000310357.3
ENST00000368549.3 ENST00000530250.1 |
GPRC6A
|
G protein-coupled receptor, family C, group 6, member A |
| chr2_-_96701722 | 0.12 |
ENST00000434632.1
|
GPAT2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr12_-_62586543 | 0.12 |
ENST00000416284.3
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chr4_-_23891658 | 0.12 |
ENST00000507380.1
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr1_+_203734296 | 0.11 |
ENST00000442561.2
ENST00000367217.5 |
LAX1
|
lymphocyte transmembrane adaptor 1 |
| chr6_+_45296391 | 0.11 |
ENST00000371436.6
ENST00000576263.1 |
RUNX2
|
runt-related transcription factor 2 |
| chr2_-_176948641 | 0.11 |
ENST00000308618.4
|
EVX2
|
even-skipped homeobox 2 |
| chr6_+_53976235 | 0.11 |
ENST00000502396.1
ENST00000358276.5 |
MLIP
|
muscular LMNA-interacting protein |
| chr4_+_24797085 | 0.11 |
ENST00000382120.3
|
SOD3
|
superoxide dismutase 3, extracellular |
| chr5_-_158526756 | 0.11 |
ENST00000313708.6
ENST00000517373.1 |
EBF1
|
early B-cell factor 1 |
| chr8_-_114449112 | 0.11 |
ENST00000455883.2
ENST00000352409.3 ENST00000297405.5 |
CSMD3
|
CUB and Sushi multiple domains 3 |
| chr3_+_153202284 | 0.11 |
ENST00000446603.2
|
C3orf79
|
chromosome 3 open reading frame 79 |
| chr20_-_23066953 | 0.11 |
ENST00000246006.4
|
CD93
|
CD93 molecule |
| chr4_-_185395672 | 0.10 |
ENST00000393593.3
|
IRF2
|
interferon regulatory factor 2 |
| chr6_-_42419649 | 0.10 |
ENST00000372922.4
ENST00000541110.1 ENST00000372917.4 |
TRERF1
|
transcriptional regulating factor 1 |
| chr12_+_18891045 | 0.10 |
ENST00000317658.3
|
CAPZA3
|
capping protein (actin filament) muscle Z-line, alpha 3 |
| chr1_+_155107820 | 0.10 |
ENST00000484157.1
|
SLC50A1
|
solute carrier family 50 (sugar efflux transporter), member 1 |
| chr3_-_50540854 | 0.10 |
ENST00000423994.2
ENST00000424201.2 ENST00000479441.1 ENST00000429770.1 |
CACNA2D2
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2 |
| chr3_-_112564797 | 0.10 |
ENST00000398214.1
ENST00000448932.1 |
CD200R1L
|
CD200 receptor 1-like |
| chr16_-_2059797 | 0.10 |
ENST00000563630.1
|
ZNF598
|
zinc finger protein 598 |
| chrX_-_54070607 | 0.10 |
ENST00000338154.6
ENST00000338946.6 |
PHF8
|
PHD finger protein 8 |
| chr10_+_21823243 | 0.10 |
ENST00000307729.7
ENST00000377091.2 |
MLLT10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr5_+_180682720 | 0.10 |
ENST00000599439.1
|
AC008443.1
|
CDNA: FLJ23158 fis, clone LNG09623; Uncharacterized protein |
| chr6_+_53976211 | 0.10 |
ENST00000503951.1
|
MLIP
|
muscular LMNA-interacting protein |
| chr18_-_22932080 | 0.10 |
ENST00000584787.1
ENST00000361524.3 ENST00000538137.2 |
ZNF521
|
zinc finger protein 521 |
| chr6_+_26045603 | 0.10 |
ENST00000540144.1
|
HIST1H3C
|
histone cluster 1, H3c |
| chr21_-_40033618 | 0.10 |
ENST00000417133.2
ENST00000398910.1 ENST00000442448.1 |
ERG
|
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chr4_-_186732241 | 0.10 |
ENST00000421639.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr1_-_114414316 | 0.10 |
ENST00000528414.1
ENST00000538253.1 ENST00000460620.1 ENST00000420377.2 ENST00000525799.1 ENST00000359785.5 |
PTPN22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr7_-_135194822 | 0.10 |
ENST00000428680.2
ENST00000315544.5 ENST00000423368.2 ENST00000451834.1 ENST00000361528.4 ENST00000356162.4 ENST00000541284.1 |
CNOT4
|
CCR4-NOT transcription complex, subunit 4 |
| chr8_+_85618155 | 0.09 |
ENST00000523850.1
ENST00000521376.1 |
RALYL
|
RALY RNA binding protein-like |
| chr17_-_39646116 | 0.09 |
ENST00000328119.6
|
KRT36
|
keratin 36 |
| chr6_-_117747015 | 0.09 |
ENST00000368508.3
ENST00000368507.3 |
ROS1
|
c-ros oncogene 1 , receptor tyrosine kinase |
| chr7_-_135612198 | 0.09 |
ENST00000589735.1
|
LUZP6
|
leucine zipper protein 6 |
| chr6_+_53976285 | 0.09 |
ENST00000514433.1
|
MLIP
|
muscular LMNA-interacting protein |
| chr12_-_18890940 | 0.09 |
ENST00000543242.1
ENST00000539072.1 ENST00000541966.1 ENST00000266505.7 ENST00000447925.2 ENST00000435379.1 |
PLCZ1
|
phospholipase C, zeta 1 |
| chr3_+_109128961 | 0.09 |
ENST00000489670.1
|
RP11-702L6.4
|
RP11-702L6.4 |
| chr20_+_51588873 | 0.09 |
ENST00000371497.5
|
TSHZ2
|
teashirt zinc finger homeobox 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of DBP
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1904640 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) response to methionine(GO:1904640) |
| 0.2 | 0.6 | GO:0021919 | BMP signaling pathway involved in spinal cord dorsal/ventral patterning(GO:0021919) |
| 0.2 | 0.6 | GO:1904237 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.2 | 0.5 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.2 | 0.5 | GO:0021569 | rhombomere 3 development(GO:0021569) rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.2 | 0.8 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 0.6 | GO:1904799 | negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.1 | 0.4 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.4 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.1 | 0.6 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.4 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.1 | 0.5 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 0.1 | GO:0060029 | convergent extension involved in organogenesis(GO:0060029) |
| 0.1 | 0.3 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.1 | 0.3 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 0.4 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.1 | 0.4 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 0.6 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.1 | 0.2 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.1 | 0.2 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.1 | 0.4 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 0.3 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 0.4 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 0.2 | GO:0060913 | cardiac cell fate determination(GO:0060913) |
| 0.0 | 0.2 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.2 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.5 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.0 | 0.9 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.3 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.5 | GO:1902961 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.1 | GO:0021546 | rhombomere development(GO:0021546) |
| 0.0 | 0.1 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.0 | 0.7 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.2 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.3 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.1 | GO:0070425 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.1 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.3 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.1 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.0 | 0.3 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.4 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.1 | GO:1900155 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.0 | 0.0 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.4 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.3 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.1 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.2 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.1 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.0 | GO:1905069 | allantois development(GO:1905069) |
| 0.0 | 1.8 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.6 | GO:0006882 | cellular zinc ion homeostasis(GO:0006882) zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.0 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) regulation of positive thymic T cell selection(GO:1902232) |
| 0.0 | 0.4 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.0 | GO:0090177 | establishment of planar polarity involved in neural tube closure(GO:0090177) |
| 0.0 | 0.3 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.5 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.7 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.1 | GO:1901545 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.0 | 0.1 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.8 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.4 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.2 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.0 | 0.9 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.6 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) polyamine binding(GO:0019808) |
| 0.1 | 0.5 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 0.4 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.1 | 0.3 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.6 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.3 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 0.2 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.1 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.0 | 0.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 1.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.4 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.6 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.2 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.4 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.4 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.2 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.1 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.4 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.2 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.5 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.8 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.8 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 1.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.4 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.6 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |