Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
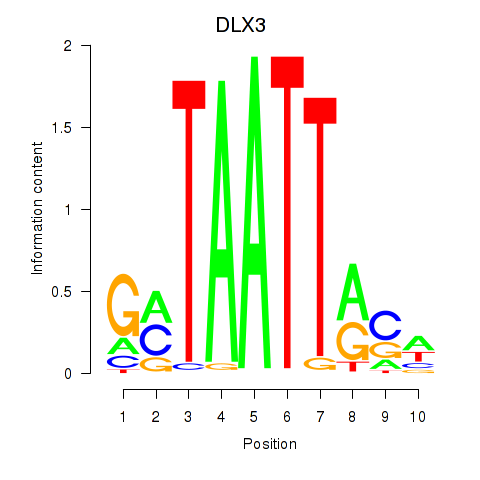
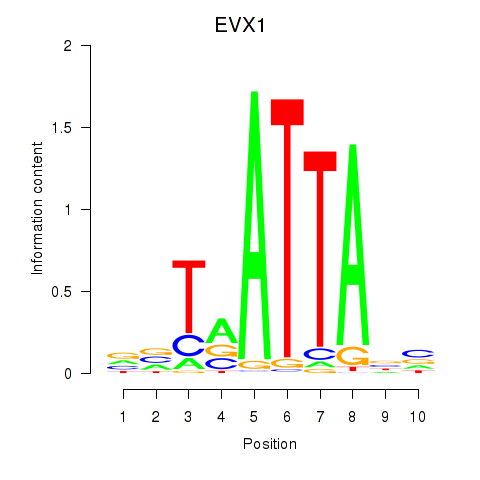
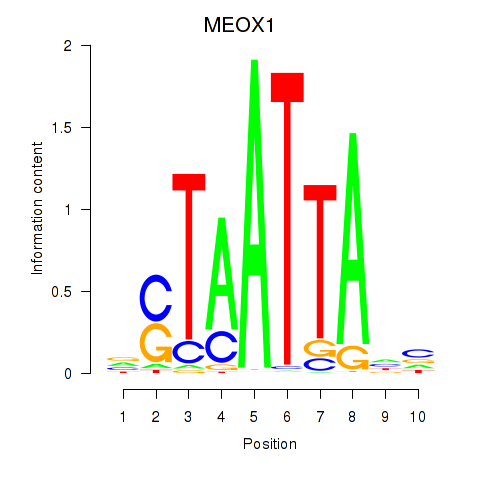
Results for DLX3_EVX1_MEOX1
Z-value: 0.44
Transcription factors associated with DLX3_EVX1_MEOX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DLX3
|
ENSG00000064195.7 | distal-less homeobox 3 |
|
EVX1
|
ENSG00000106038.8 | even-skipped homeobox 1 |
|
MEOX1
|
ENSG00000005102.8 | mesenchyme homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DLX3 | hg19_v2_chr17_-_48072574_48072597 | -0.69 | 7.5e-04 | Click! |
| MEOX1 | hg19_v2_chr17_-_41739283_41739322 | -0.32 | 1.7e-01 | Click! |
| EVX1 | hg19_v2_chr7_+_27282319_27282394 | 0.17 | 4.8e-01 | Click! |
Activity profile of DLX3_EVX1_MEOX1 motif
Sorted Z-values of DLX3_EVX1_MEOX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_59910132 | 1.52 |
ENST00000559200.1
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr7_-_14029283 | 1.18 |
ENST00000433547.1
ENST00000405192.2 |
ETV1
|
ets variant 1 |
| chr6_+_151646800 | 1.16 |
ENST00000354675.6
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr9_+_116225999 | 0.97 |
ENST00000317613.6
|
RGS3
|
regulator of G-protein signaling 3 |
| chr15_+_80351910 | 0.94 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr15_-_64673630 | 0.92 |
ENST00000558008.1
ENST00000559519.1 ENST00000380258.2 |
KIAA0101
|
KIAA0101 |
| chr17_-_33446735 | 0.88 |
ENST00000460118.2
ENST00000335858.7 |
RAD51D
|
RAD51 paralog D |
| chr20_+_60174827 | 0.85 |
ENST00000543233.1
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal) |
| chr17_-_33446820 | 0.82 |
ENST00000592577.1
ENST00000590016.1 ENST00000345365.6 ENST00000360276.3 ENST00000357906.3 |
RAD51D
|
RAD51 paralog D |
| chr17_-_46690839 | 0.81 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr15_-_64673665 | 0.78 |
ENST00000300035.4
|
KIAA0101
|
KIAA0101 |
| chr16_-_29910853 | 0.74 |
ENST00000308713.5
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
| chr6_-_76203454 | 0.73 |
ENST00000237172.7
|
FILIP1
|
filamin A interacting protein 1 |
| chr7_-_99716940 | 0.73 |
ENST00000440225.1
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr11_-_31531121 | 0.73 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr7_+_6655225 | 0.71 |
ENST00000457543.3
|
ZNF853
|
zinc finger protein 853 |
| chr6_-_76203345 | 0.70 |
ENST00000393004.2
|
FILIP1
|
filamin A interacting protein 1 |
| chr3_+_44840679 | 0.69 |
ENST00000425755.1
|
KIF15
|
kinesin family member 15 |
| chr17_-_57229155 | 0.67 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr17_-_46716647 | 0.63 |
ENST00000608940.1
|
RP11-357H14.17
|
RP11-357H14.17 |
| chr1_-_21625486 | 0.62 |
ENST00000481130.2
|
ECE1
|
endothelin converting enzyme 1 |
| chr5_-_159827033 | 0.62 |
ENST00000523213.1
|
C5orf54
|
chromosome 5 open reading frame 54 |
| chr16_+_53133070 | 0.62 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr8_+_40010989 | 0.61 |
ENST00000315792.3
|
C8orf4
|
chromosome 8 open reading frame 4 |
| chr10_-_92681033 | 0.60 |
ENST00000371697.3
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr7_-_32338917 | 0.58 |
ENST00000396193.1
|
PDE1C
|
phosphodiesterase 1C, calmodulin-dependent 70kDa |
| chr20_+_43029911 | 0.58 |
ENST00000443598.2
ENST00000316099.4 ENST00000415691.2 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr11_+_47293795 | 0.57 |
ENST00000422579.1
|
MADD
|
MAP-kinase activating death domain |
| chr4_-_139163491 | 0.57 |
ENST00000280612.5
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr7_-_33102399 | 0.57 |
ENST00000242210.7
|
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr7_-_33102338 | 0.56 |
ENST00000610140.1
|
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr12_+_107712173 | 0.56 |
ENST00000280758.5
ENST00000420571.2 |
BTBD11
|
BTB (POZ) domain containing 11 |
| chrX_-_106243294 | 0.54 |
ENST00000255495.7
|
MORC4
|
MORC family CW-type zinc finger 4 |
| chr5_+_174151536 | 0.54 |
ENST00000239243.6
ENST00000507785.1 |
MSX2
|
msh homeobox 2 |
| chr15_-_37393406 | 0.54 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr15_+_89631647 | 0.53 |
ENST00000569550.1
ENST00000565066.1 ENST00000565973.1 |
ABHD2
|
abhydrolase domain containing 2 |
| chr15_-_55562582 | 0.53 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr7_-_99716952 | 0.52 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr15_+_89631381 | 0.52 |
ENST00000352732.5
|
ABHD2
|
abhydrolase domain containing 2 |
| chr14_-_95236551 | 0.51 |
ENST00000238558.3
|
GSC
|
goosecoid homeobox |
| chr5_+_156712372 | 0.51 |
ENST00000541131.1
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr12_-_120241187 | 0.49 |
ENST00000392520.2
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr12_-_56694083 | 0.49 |
ENST00000552688.1
ENST00000548041.1 ENST00000551137.1 ENST00000551968.1 ENST00000542324.2 ENST00000546930.1 ENST00000549221.1 ENST00000550159.1 ENST00000550734.1 |
CS
|
citrate synthase |
| chr6_+_126221034 | 0.48 |
ENST00000433571.1
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr1_-_197115818 | 0.48 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr1_-_232651312 | 0.48 |
ENST00000262861.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chr12_+_78359999 | 0.47 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr15_-_55563072 | 0.47 |
ENST00000567380.1
ENST00000565972.1 ENST00000569493.1 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr14_-_37051798 | 0.46 |
ENST00000258829.5
|
NKX2-8
|
NK2 homeobox 8 |
| chr1_-_159832438 | 0.46 |
ENST00000368100.1
|
VSIG8
|
V-set and immunoglobulin domain containing 8 |
| chr16_+_57844549 | 0.46 |
ENST00000564282.1
|
CTD-2600O9.1
|
uncharacterized protein LOC388282 |
| chr17_-_64225508 | 0.45 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr17_+_43238438 | 0.45 |
ENST00000593138.1
ENST00000586681.1 |
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chrX_-_106243451 | 0.44 |
ENST00000355610.4
ENST00000535534.1 |
MORC4
|
MORC family CW-type zinc finger 4 |
| chr4_+_105828537 | 0.43 |
ENST00000515649.1
|
RP11-556I14.1
|
RP11-556I14.1 |
| chr11_-_117747327 | 0.42 |
ENST00000584230.1
ENST00000527429.1 ENST00000584394.1 ENST00000532984.1 |
FXYD6
FXYD6-FXYD2
|
FXYD domain containing ion transport regulator 6 FXYD6-FXYD2 readthrough |
| chr2_-_61697862 | 0.41 |
ENST00000398571.2
|
USP34
|
ubiquitin specific peptidase 34 |
| chr14_-_57960545 | 0.41 |
ENST00000526336.1
ENST00000216445.3 |
C14orf105
|
chromosome 14 open reading frame 105 |
| chr4_+_41614909 | 0.41 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr14_-_57960456 | 0.41 |
ENST00000534126.1
ENST00000422976.2 |
C14orf105
|
chromosome 14 open reading frame 105 |
| chr7_-_14029515 | 0.41 |
ENST00000430479.1
ENST00000405218.2 ENST00000343495.5 |
ETV1
|
ets variant 1 |
| chr12_+_28410128 | 0.41 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr8_+_22424551 | 0.41 |
ENST00000523348.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr7_-_111424506 | 0.40 |
ENST00000450156.1
ENST00000494651.2 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr20_-_50419055 | 0.40 |
ENST00000217086.4
|
SALL4
|
spalt-like transcription factor 4 |
| chr8_-_19614810 | 0.39 |
ENST00000524213.1
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr6_+_142623758 | 0.39 |
ENST00000541199.1
ENST00000435011.2 |
GPR126
|
G protein-coupled receptor 126 |
| chr13_+_78315466 | 0.39 |
ENST00000314070.5
ENST00000462234.1 |
SLAIN1
|
SLAIN motif family, member 1 |
| chr14_+_67831576 | 0.39 |
ENST00000555876.1
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
| chrX_+_10031499 | 0.37 |
ENST00000454666.1
|
WWC3
|
WWC family member 3 |
| chr5_+_159343688 | 0.37 |
ENST00000306675.3
|
ADRA1B
|
adrenoceptor alpha 1B |
| chr20_+_42187682 | 0.36 |
ENST00000373092.3
ENST00000373077.1 |
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr12_-_56693758 | 0.36 |
ENST00000547298.1
ENST00000551936.1 ENST00000551253.1 ENST00000551473.1 |
CS
|
citrate synthase |
| chr15_-_65903407 | 0.35 |
ENST00000395644.4
ENST00000567744.1 ENST00000568573.1 ENST00000562830.1 ENST00000569491.1 ENST00000561769.1 |
VWA9
|
von Willebrand factor A domain containing 9 |
| chr8_+_32579271 | 0.34 |
ENST00000518084.1
|
NRG1
|
neuregulin 1 |
| chr6_+_105404899 | 0.33 |
ENST00000345080.4
|
LIN28B
|
lin-28 homolog B (C. elegans) |
| chr7_-_99717463 | 0.33 |
ENST00000437822.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr4_+_41614720 | 0.32 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr8_+_32579341 | 0.32 |
ENST00000519240.1
ENST00000539990.1 |
NRG1
|
neuregulin 1 |
| chr8_+_32579321 | 0.31 |
ENST00000522402.1
|
NRG1
|
neuregulin 1 |
| chr15_+_89164560 | 0.31 |
ENST00000379231.3
ENST00000559528.1 |
AEN
|
apoptosis enhancing nuclease |
| chr22_-_30960876 | 0.31 |
ENST00000401975.1
ENST00000428682.1 ENST00000423299.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr12_+_54378849 | 0.30 |
ENST00000515593.1
|
HOXC10
|
homeobox C10 |
| chr10_-_33623310 | 0.29 |
ENST00000395995.1
ENST00000374823.5 ENST00000374821.5 ENST00000374816.3 |
NRP1
|
neuropilin 1 |
| chr10_-_27529486 | 0.29 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr20_+_43990576 | 0.29 |
ENST00000372727.1
ENST00000414310.1 |
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr7_-_111424462 | 0.29 |
ENST00000437129.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr7_-_14028488 | 0.29 |
ENST00000405358.4
|
ETV1
|
ets variant 1 |
| chr12_+_54378923 | 0.28 |
ENST00000303460.4
|
HOXC10
|
homeobox C10 |
| chr6_-_116833500 | 0.28 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr13_-_31191642 | 0.28 |
ENST00000405805.1
|
HMGB1
|
high mobility group box 1 |
| chr10_+_35484793 | 0.28 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr13_-_30160925 | 0.28 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr19_+_34287751 | 0.28 |
ENST00000590771.1
ENST00000589786.1 ENST00000284006.6 ENST00000588881.1 |
KCTD15
|
potassium channel tetramerization domain containing 15 |
| chr3_-_123339343 | 0.27 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr20_+_3776371 | 0.27 |
ENST00000245960.5
|
CDC25B
|
cell division cycle 25B |
| chr16_+_57139933 | 0.27 |
ENST00000566259.1
|
CPNE2
|
copine II |
| chrX_-_19689106 | 0.26 |
ENST00000379716.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr10_-_101690650 | 0.26 |
ENST00000543621.1
|
DNMBP
|
dynamin binding protein |
| chr20_+_42187608 | 0.26 |
ENST00000373100.1
|
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr3_-_33686925 | 0.25 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr13_+_36050881 | 0.24 |
ENST00000537702.1
|
NBEA
|
neurobeachin |
| chr17_+_73539339 | 0.24 |
ENST00000581713.1
|
LLGL2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr5_+_137722255 | 0.24 |
ENST00000542866.1
|
KDM3B
|
lysine (K)-specific demethylase 3B |
| chr18_+_32556892 | 0.24 |
ENST00000591734.1
ENST00000413393.1 ENST00000589180.1 ENST00000587359.1 |
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr15_+_80351977 | 0.23 |
ENST00000559157.1
ENST00000561012.1 ENST00000564367.1 ENST00000558494.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr6_+_52535878 | 0.23 |
ENST00000211314.4
|
TMEM14A
|
transmembrane protein 14A |
| chr8_-_17533838 | 0.23 |
ENST00000400046.1
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr17_+_42264556 | 0.23 |
ENST00000319511.6
ENST00000589785.1 ENST00000592825.1 ENST00000589184.1 |
TMUB2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr2_-_55647057 | 0.23 |
ENST00000436346.1
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr5_-_142780280 | 0.22 |
ENST00000424646.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr4_-_105416039 | 0.22 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr4_+_66536248 | 0.22 |
ENST00000514260.1
ENST00000507117.1 |
RP11-807H7.1
|
RP11-807H7.1 |
| chr12_-_15815626 | 0.22 |
ENST00000540613.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr13_+_78315528 | 0.21 |
ENST00000496045.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr2_-_55646957 | 0.21 |
ENST00000263630.8
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr1_+_202317855 | 0.20 |
ENST00000356764.2
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr15_-_55562479 | 0.20 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr2_+_113299990 | 0.20 |
ENST00000537335.1
ENST00000417433.2 |
POLR1B
|
polymerase (RNA) I polypeptide B, 128kDa |
| chr4_-_174451370 | 0.20 |
ENST00000359562.4
|
HAND2
|
heart and neural crest derivatives expressed 2 |
| chr17_-_48785216 | 0.20 |
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr7_+_116660246 | 0.20 |
ENST00000434836.1
ENST00000393443.1 ENST00000465133.1 ENST00000477742.1 ENST00000393447.4 ENST00000393444.3 |
ST7
|
suppression of tumorigenicity 7 |
| chr11_+_101918153 | 0.20 |
ENST00000434758.2
ENST00000526781.1 ENST00000534360.1 |
C11orf70
|
chromosome 11 open reading frame 70 |
| chr12_-_56694142 | 0.19 |
ENST00000550655.1
ENST00000548567.1 ENST00000551430.2 ENST00000351328.3 |
CS
|
citrate synthase |
| chr15_+_75080883 | 0.19 |
ENST00000567571.1
|
CSK
|
c-src tyrosine kinase |
| chr20_-_50418972 | 0.19 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr8_-_1922789 | 0.19 |
ENST00000521498.1
|
RP11-439C15.4
|
RP11-439C15.4 |
| chr11_-_124981475 | 0.19 |
ENST00000532156.1
ENST00000532407.1 ENST00000279968.4 ENST00000527766.1 ENST00000529583.1 ENST00000524373.1 ENST00000527271.1 ENST00000526175.1 ENST00000529609.1 ENST00000533273.1 ENST00000531909.1 ENST00000529530.1 |
TMEM218
|
transmembrane protein 218 |
| chr4_-_155533787 | 0.19 |
ENST00000407946.1
ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG
|
fibrinogen gamma chain |
| chr1_-_190446759 | 0.19 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr16_-_66864806 | 0.19 |
ENST00000566336.1
ENST00000394074.2 ENST00000563185.2 ENST00000359087.4 ENST00000379463.2 ENST00000565535.1 ENST00000290810.3 |
NAE1
|
NEDD8 activating enzyme E1 subunit 1 |
| chr6_-_76072719 | 0.18 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chrX_-_46187069 | 0.18 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr16_-_69390209 | 0.18 |
ENST00000563634.1
|
RP11-343C2.9
|
Uncharacterized protein |
| chr4_-_66536057 | 0.18 |
ENST00000273854.3
|
EPHA5
|
EPH receptor A5 |
| chr3_-_33686743 | 0.18 |
ENST00000333778.6
ENST00000539981.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr6_-_111804905 | 0.18 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr14_+_74034310 | 0.18 |
ENST00000538782.1
|
ACOT2
|
acyl-CoA thioesterase 2 |
| chr18_-_44181442 | 0.18 |
ENST00000398722.4
|
LOXHD1
|
lipoxygenase homology domains 1 |
| chr13_+_78315295 | 0.18 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr2_-_37544209 | 0.18 |
ENST00000234179.2
|
PRKD3
|
protein kinase D3 |
| chr4_-_66536196 | 0.17 |
ENST00000511294.1
|
EPHA5
|
EPH receptor A5 |
| chr4_-_138453606 | 0.17 |
ENST00000412923.2
ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18
|
protocadherin 18 |
| chr6_-_75994025 | 0.17 |
ENST00000518161.1
|
TMEM30A
|
transmembrane protein 30A |
| chr3_+_159557637 | 0.17 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr4_+_95916947 | 0.17 |
ENST00000506363.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr17_-_2117600 | 0.17 |
ENST00000572369.1
|
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr6_-_111136299 | 0.17 |
ENST00000457688.1
|
CDK19
|
cyclin-dependent kinase 19 |
| chr4_+_105828492 | 0.16 |
ENST00000506148.1
|
RP11-556I14.1
|
RP11-556I14.1 |
| chr2_-_17981462 | 0.16 |
ENST00000402989.1
ENST00000428868.1 |
SMC6
|
structural maintenance of chromosomes 6 |
| chr5_-_87516448 | 0.16 |
ENST00000511218.1
|
TMEM161B
|
transmembrane protein 161B |
| chrX_-_72434628 | 0.16 |
ENST00000536638.1
ENST00000373517.3 |
NAP1L2
|
nucleosome assembly protein 1-like 2 |
| chr2_-_224467002 | 0.16 |
ENST00000421386.1
ENST00000433889.1 |
SCG2
|
secretogranin II |
| chr13_-_45010939 | 0.16 |
ENST00000261489.2
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr6_+_127898312 | 0.16 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr12_-_53171128 | 0.15 |
ENST00000332411.2
|
KRT76
|
keratin 76 |
| chr10_-_21186144 | 0.15 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr14_-_51027838 | 0.15 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr8_+_110346546 | 0.15 |
ENST00000521662.1
ENST00000521688.1 ENST00000520147.1 |
ENY2
|
enhancer of yellow 2 homolog (Drosophila) |
| chr20_-_271304 | 0.15 |
ENST00000400269.3
ENST00000360321.2 |
C20orf96
|
chromosome 20 open reading frame 96 |
| chr19_-_45681482 | 0.15 |
ENST00000592647.1
ENST00000006275.4 ENST00000588062.1 ENST00000585934.1 |
TRAPPC6A
|
trafficking protein particle complex 6A |
| chr20_+_18447771 | 0.15 |
ENST00000377603.4
|
POLR3F
|
polymerase (RNA) III (DNA directed) polypeptide F, 39 kDa |
| chr8_-_101963482 | 0.15 |
ENST00000419477.2
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr2_-_86422095 | 0.15 |
ENST00000254636.5
|
IMMT
|
inner membrane protein, mitochondrial |
| chr3_-_57233966 | 0.14 |
ENST00000473921.1
ENST00000295934.3 |
HESX1
|
HESX homeobox 1 |
| chr14_+_56584414 | 0.14 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr17_-_71223839 | 0.14 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr6_+_4087664 | 0.14 |
ENST00000430835.2
|
C6orf201
|
chromosome 6 open reading frame 201 |
| chr9_+_130026756 | 0.13 |
ENST00000314904.5
ENST00000373387.4 |
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr5_-_16509101 | 0.13 |
ENST00000399793.2
|
FAM134B
|
family with sequence similarity 134, member B |
| chr17_-_1418972 | 0.13 |
ENST00000571274.1
|
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr15_+_89164520 | 0.13 |
ENST00000332810.3
|
AEN
|
apoptosis enhancing nuclease |
| chr11_-_47546220 | 0.13 |
ENST00000528538.1
|
CELF1
|
CUGBP, Elav-like family member 1 |
| chr8_-_110346614 | 0.13 |
ENST00000239690.4
|
NUDCD1
|
NudC domain containing 1 |
| chr18_-_56985873 | 0.13 |
ENST00000299721.3
|
CPLX4
|
complexin 4 |
| chr12_+_64798826 | 0.13 |
ENST00000540203.1
|
XPOT
|
exportin, tRNA |
| chr1_+_101003687 | 0.13 |
ENST00000315033.4
|
GPR88
|
G protein-coupled receptor 88 |
| chr6_-_111136513 | 0.13 |
ENST00000368911.3
|
CDK19
|
cyclin-dependent kinase 19 |
| chr9_-_34329198 | 0.13 |
ENST00000379166.2
ENST00000345050.2 |
KIF24
|
kinesin family member 24 |
| chr1_-_219615984 | 0.13 |
ENST00000420762.1
|
RP11-95P13.1
|
RP11-95P13.1 |
| chr4_+_146402346 | 0.12 |
ENST00000514778.1
ENST00000507594.1 |
SMAD1
|
SMAD family member 1 |
| chr17_+_44679808 | 0.12 |
ENST00000571172.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr12_-_10978957 | 0.12 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr17_-_38956205 | 0.12 |
ENST00000306658.7
|
KRT28
|
keratin 28 |
| chr13_+_78315348 | 0.12 |
ENST00000441784.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr7_+_99717230 | 0.12 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr13_-_28545276 | 0.12 |
ENST00000381020.7
|
CDX2
|
caudal type homeobox 2 |
| chr1_-_243326612 | 0.12 |
ENST00000492145.1
ENST00000490813.1 ENST00000464936.1 |
CEP170
|
centrosomal protein 170kDa |
| chr9_+_82188077 | 0.12 |
ENST00000425506.1
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr2_-_27498208 | 0.11 |
ENST00000424577.1
ENST00000426569.1 |
SLC30A3
|
solute carrier family 30 (zinc transporter), member 3 |
| chr13_+_110958124 | 0.11 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr20_-_50418947 | 0.11 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr9_-_136933134 | 0.11 |
ENST00000303407.7
|
BRD3
|
bromodomain containing 3 |
| chr8_-_101963677 | 0.11 |
ENST00000395956.3
ENST00000395953.2 |
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr11_-_117747434 | 0.11 |
ENST00000529335.2
ENST00000530956.1 ENST00000260282.4 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr2_-_99279928 | 0.11 |
ENST00000414521.2
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr5_-_148929848 | 0.11 |
ENST00000504676.1
ENST00000515435.1 |
CSNK1A1
|
casein kinase 1, alpha 1 |
| chr2_+_201754135 | 0.11 |
ENST00000409357.1
ENST00000409129.2 |
NIF3L1
|
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr1_+_236958554 | 0.10 |
ENST00000366577.5
ENST00000418145.2 |
MTR
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr11_-_95657231 | 0.10 |
ENST00000409459.1
ENST00000352297.7 ENST00000393223.3 ENST00000346299.5 |
MTMR2
|
myotubularin related protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of DLX3_EVX1_MEOX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.2 | 1.7 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.2 | 0.5 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.2 | 0.5 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 1.0 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 1.8 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 0.4 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.1 | 0.4 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 1.0 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 0.6 | GO:0034959 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.1 | 0.4 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.1 | 0.7 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 0.7 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.3 | GO:0035711 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) |
| 0.1 | 0.6 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 2.1 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 1.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.3 | GO:1904835 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.1 | 0.6 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 0.6 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.4 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 | 0.5 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.7 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.4 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.3 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.3 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.1 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.3 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 1.2 | GO:0006625 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.5 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.2 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 1.0 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.1 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.4 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.4 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.4 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.2 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.3 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.2 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.2 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.1 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.8 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.0 | 0.6 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.1 | GO:2001153 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.2 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 1.0 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.5 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.2 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.2 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.3 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.4 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.1 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.2 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.1 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.0 | 0.3 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.1 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.1 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) bone marrow development(GO:0048539) |
| 0.0 | 0.5 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.1 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.3 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.2 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.1 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.0 | 0.1 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.1 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.2 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.1 | GO:0007019 | microtubule depolymerization(GO:0007019) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.2 | 1.7 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.2 | 0.7 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.1 | 0.4 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) glial limiting end-foot(GO:0097451) |
| 0.1 | 0.6 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.1 | 1.0 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.9 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 1.6 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.6 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.5 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.7 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.4 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.3 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.2 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.4 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 1.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.1 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.3 | 1.0 | GO:0036440 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.2 | 1.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.2 | 0.6 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.7 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.1 | 0.4 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 1.7 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 1.6 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.4 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 0.1 | 1.0 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 0.2 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.1 | 1.0 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.6 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.1 | 0.4 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.0 | 1.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.3 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.1 | GO:0015633 | zinc transporting ATPase activity(GO:0015633) |
| 0.0 | 0.4 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.3 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 1.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.2 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.1 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.3 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.4 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.6 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.1 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.0 | GO:0035877 | death effector domain binding(GO:0035877) cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 1.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.0 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.1 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |